Cotyledon Node VIGS Delivery in Soybean: An Optimized Protocol for Rapid Gene Function Discovery
This article provides a comprehensive resource for researchers and scientists on the implementation and optimization of Virus-Induced Gene Silencing (VIGS) in soybean via the cotyledon node delivery method.
Cotyledon Node VIGS Delivery in Soybean: An Optimized Protocol for Rapid Gene Function Discovery
Abstract
This article provides a comprehensive resource for researchers and scientists on the implementation and optimization of Virus-Induced Gene Silencing (VIGS) in soybean via the cotyledon node delivery method. We cover foundational principles of VIGS as a rapid alternative to stable transformation, detail a step-by-step methodological protocol for Agrobacterium-mediated cotyledon node infection, and present key troubleshooting and optimization strategies to achieve high silencing efficiency. Furthermore, we validate the system's robustness through case studies of resistance gene discovery and provide a comparative analysis with other soybean VIGS vectors. This guide aims to empower functional genomics and drug discovery research by facilitating high-throughput gene function validation in soybean.
Understanding VIGS and the Cotyledon Node Advantage in Soybean Functional Genomics
The Critical Need for Rapid Gene Validation in Soybean
Soybean ( Glycine max L.) is a vital global crop, serving as a primary source of plant-based protein and oil for food and feed. However, its productivity is consistently threatened by biotic and abiotic stresses, including diseases and heat stress, which can cause severe yield losses [1] [2]. The development of resilient cultivars represents the most sustainable strategy to mitigate these losses. A major bottleneck in this process is the functional characterization of candidate genes discovered through genomic studies. Stable genetic transformation, a common approach for functional analysis, is notoriously time-consuming and labor-intensive in soybean [1]. Virus-Induced Gene Silencing (VIGS) has emerged as a powerful alternative, enabling rapid in planta assessment of gene function. This application note details an optimized, highly efficient Tobacco Rattle Virus (TRV)-based VIGS protocol for soybean, utilizing Agrobacterium tumefaciens-mediated delivery via the cotyledon node. This method achieves systemic silencing with an efficiency of 65% to 95%, facilitating rapid phenotypic validation and accelerating soybean breeding programs [1].
Technical Specifications and Performance Metrics
The established TRV-VIGS system demonstrates high efficiency in silencing endogenous genes, leading to discernible phenotypic changes. The table below summarizes key quantitative data from validation experiments.
Table 1: Performance Metrics of the TRV-VIGS System in Soybean
| Parameter / Gene Target | Measurement / Phenotype | Silencing Efficiency | Key Findings |
|---|---|---|---|
| Agroinfection Efficiency | >80% of cells in transverse section (cv. Tianlong 1) | N/A | Fluorescence signal confirmed successful T-DNA delivery [1] |
| GmPDS | Photobleaching in leaves and cluster buds | High | Phenotype observed at 21 days post-inoculation (dpi) [1] |
| GmRpp6907 | Compromised rust resistance | 65% - 95% | Confirmed system's utility for disease resistance validation [1] |
| GmRPT4 | Altered defense response | 65% - 95% | Demonstrated role in defense mechanisms [1] |
Materials and Reagents
Research Reagent Solutions
The following table lists the essential materials required for the execution of the cotyledon node VIGS protocol.
Table 2: Key Research Reagents and Their Functions in the VIGS Protocol
| Reagent / Material | Function / Application | Specifications / Notes |
|---|---|---|
| pTRV1 and pTRV2 Vectors | TRV-based VIGS vector system | pTRV2 derivatives carry target gene fragments for silencing [1] |
| Agrobacterium tumefaciens GV3101 | Delivery vehicle for TRV vectors | Host strain for plasmid transformation and plant infection [1] |
| Soybean Cultivars | Plant material for VIGS | e.g., Tianlong 1; requires optimization for different genotypes [1] |
| Restriction Enzymes (EcoRI, XhoI) | Molecular cloning | Used for digestion of pTRV2 vector and insertion of target gene fragment [1] |
Step-by-Step Protocol
Vector Construction and Agrobacterium Preparation
- Step 1: Clone Target Gene Fragment. Amplify a ~200-500 bp fragment of the target gene (e.g., GmPDS) from soybean cDNA using gene-specific primers with engineered EcoRI and XhoI restriction sites [1]. Primer sequences for GmPDS are: PDS-F: 5'-taaggttaccGAATTCTCTCCGCGTCCTCTAAAAC-3' and PDS-R: 5'-atgcccgggcCTCGAGTCCAGGCTTATTTGGCATAGC-3' [1].
- Step 2: Ligate and Transform. Digest the pTRV2-GFP vector with EcoRI and XhoI. Ligate the purified PCR product into the linearized vector. Transform the ligation product into E. coli DH5α competent cells and select positive clones for sequence verification [1].
- Step 3: Mobilize into Agrobacterium. Introduce the confirmed recombinant plasmid and the pTRV1 plasmid into Agrobacterium tumefaciens strain GV3101 via electroporation or freeze-thaw transformation [1].
- Step 4: Prepare Agrobacterium Culture. Inoculate single colonies of Agrobacterium containing pTRV1 and pTRV2-derivatives (e.g., pTRV:empty, pTRV:GmPDS) in liquid medium with appropriate antibiotics. Grow cultures at 28°C to an OD₆₀₀ of ~0.8-1.0. Pellet the cells and resuspend in an induction medium (e.g., containing MES and Acetosyringone) to a final OD₆₀₀ of 1.0. Incubate the suspensions for 3-4 hours at room temperature [1].
Plant Material Preparation and Agroinfiltration
- Step 5: Prepare Soybean Explants. Surface-sterilize soybean seeds and soak them in sterile water until swollen. Under sterile conditions, longitudinally bisect the seeds to obtain half-seed explants, ensuring the cotyledonary node is intact and exposed [1].
- Step 6: Mix Agrobacterium Suspensions. Combine equal volumes of the induced Agrobacterium suspensions harboring pTRV1 and the recombinant pTRV2 (e.g., pTRV2-GmPDS) [1].
- Step 7: Infect Explants. Immerse the fresh half-seed explants in the mixed Agrobacterium suspension for 20-30 minutes, ensuring full contact with the cotyledonary node. This duration has been optimized for maximum infection efficiency [1].
- Step 8: Co-cultivation and Plant Regeneration. Blot-dry the explants and transfer them to co-cultivation media. Maintain the plates in the dark at 25°C for 2-3 days. Subsequently, transfer the explants to regeneration media containing antibiotics to suppress Agrobacterium overgrowth and encourage shoot development. Maintain plants under standard growth chamber conditions (e.g., 25°C, 16/8 light/dark cycle) [1].
The following diagram illustrates the complete experimental workflow from vector construction to phenotypic analysis.
Data Analysis and Interpretation
- Phenotypic Monitoring: Systemically silenced plants typically exhibit phenotypes, such as photobleaching for GmPDS, beginning at 21 days post-inoculation (dpi). Initial signs often appear in the cluster buds before spreading to newly developed leaves [1].
- Molecular Validation: Confirm silencing efficiency at the transcript level using quantitative RT-PCR (qRT-PCR). Compare mRNA levels of the target gene in pTRV:target gene plants versus pTRV:empty vector controls. A successful experiment should show a significant reduction (65-95%) in target gene expression [1].
- Infection Efficiency Check: Around 4 days post-infection, excise a portion of the hypocotyl and observe under a fluorescence microscope for GFP signal. The presence of fluorescence in over 80% of cells in a transverse section indicates high infection efficiency, which is a reliable predictor of successful silencing [1].
Discussion and Application
This optimized TRV-VIGS protocol addresses a critical bottleneck in soybean research by providing a rapid and efficient method for high-throughput gene function validation. The key advantage lies in its ability to bypass the lengthy process of stable transformation, allowing researchers to screen candidate genes in a matter of weeks. The method's high efficiency (65-95%) and systemic nature make it particularly suitable for functional studies of agronomically important traits, such as disease resistance (e.g., GmRpp6907 for rust resistance) and abiotic stress tolerance [1].
The integration of this VIGS platform with other advanced genomic tools, such as CRISPR/Cas genome editing and spatial transcriptomics, creates a powerful pipeline for soybean improvement [3] [4]. Candidate genes identified through genome-wide association studies (GWAS) or QTL mapping can be rapidly screened and validated using this VIGS system [2] [5]. Subsequently, precise modifications can be introduced via CRISPR/Cas to develop elite germplasm with enhanced resilience and yield [3] [6]. This synergistic approach significantly accelerates the breeding cycle, contributing to the overarching goal of ensuring global food security in the face of climate change.
Virus-Induced Gene Silencing (VIGS) is a powerful reverse genetics tool that harnesses the plant's innate antiviral defense mechanism for post-transcriptional gene silencing (PTGS). As a rapid and versatile alternative to stable genetic transformation, VIGS utilizes recombinant viral vectors to deliver fragments of plant genes, triggering sequence-specific degradation of complementary endogenous mRNA [7]. This technology has become indispensable for functional genomics, particularly in recalcitrant species like soybean, where traditional transformation methods are time-consuming and genotype-dependent [1] [8]. The application of VIGS in soybean research has gained significant momentum, with recent methodological advances enabling more efficient investigation of disease resistance, stress tolerance, and developmental processes [1] [9] [10].
The core principle of VIGS exploits the plant's RNA interference (RNAi) pathway. When a viral vector containing a fragment of a host gene replicates in plant cells, double-stranded RNA (dsRNA) is produced and recognized by the plant's Dicer-like enzymes (DCL). These process the dsRNA into 21- to 24-nucleotide small interfering RNAs (siRNAs), which are then incorporated into the RNA-induced silencing complex (RISC). The activated RISC complex guides sequence-specific cleavage of complementary endogenous mRNA transcripts, effectively silencing the target gene [7].
Key VIGS Vectors and Their Applications
Comparative Analysis of Viral Vectors
Various viral vectors have been engineered for VIGS, each with distinct advantages and host range specificities. The selection of an appropriate vector is critical for experimental success.
Table 1: Characteristics of Major VIGS Vectors
| Vector Type | Virus Origin | Primary Hosts | Key Advantages | Limitations |
|---|---|---|---|---|
| TRV (Tobacco Rattle Virus) | RNA virus | Solanaceae, Arabidopsis, Soybean | Broad host range, mild symptoms, efficient systemic spread [1] [7] | Bipartite genome requires two vectors |
| BPMV (Bean Pod Mottle Virus) | RNA virus | Soybean | Well-established for soybean, high reliability [1] | May cause noticeable leaf symptoms |
| CSVdV (Cowpea Severe Mosaic Virus) | DNA virus | Soybean | Effective for nodulation studies [9] | More limited host range |
| ALSV (Apple Latent Spherical Virus) | RNA virus | Soybean, Legumes | Mild symptoms, broad legume application [1] | Less extensively validated than BPMV |
TRV-Based Vectors: The Gold Standard
TRV has emerged as one of the most versatile and widely adopted VIGS systems due to its broad host range, efficient systemic movement, and mild symptomatic effects on host plants [7]. The TRV genome is bipartite, requiring two separate vectors for successful silencing: TRV1 encodes replicase proteins, movement protein, and a weak RNA interference suppressor, while TRV2 contains the coat protein gene and serves as the insertion site for target gene fragments [7]. This system has been successfully optimized for soybean through Agrobacterium tumefaciens-mediated delivery via cotyledon nodes, achieving silencing efficiencies ranging from 65% to 95% [1].
Cotyledon Node VIGS Delivery in Soybean: An Optimized Protocol
The cotyledon node delivery method represents a significant advancement in soybean VIGS technology, overcoming previous limitations associated with soybean's thick cuticle and dense leaf trichomes that impeded efficient agroinfiltration [1]. This approach exploits the vascular connectivity and meristematic activity of the cotyledon node for efficient viral spread throughout the plant.
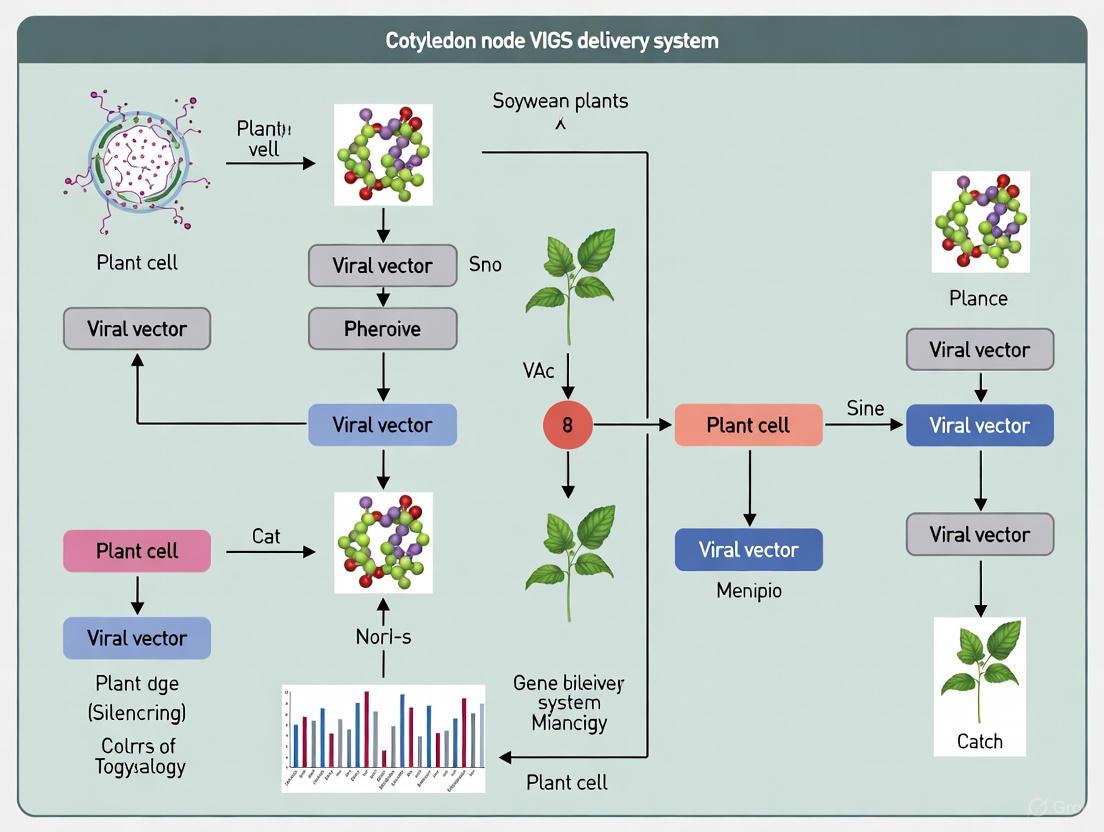
Diagram 1: Cotyledon Node VIGS Workflow for Soybean. This diagram illustrates the optimized procedural sequence for implementing VIGS in soybean via cotyledon node delivery, from seed preparation to phenotypic validation.
Detailed Experimental Protocol
Vector Construction and Agrobacterium Preparation
The first critical step involves engineering the appropriate VIGS construct and preparing the Agrobacterium strain for plant transformation.
Cloning Target Gene Fragments:
- Amplify 200-300 bp gene-specific fragments from soybean cDNA using sequence-specific primers with added restriction sites (e.g., EcoRI and XhoI) [1]
- Ligate the purified PCR product into the pTRV2 vector predigested with corresponding restriction enzymes
- Transform the ligation product into E. coli DH5α competent cells and verify positive clones by sequencing [1]
- Introduce confirmed recombinant plasmids into Agrobacterium tumefaciens strain GV3101 through electroporation or freeze-thaw method
Agrobacterium Culture Preparation:
- Inoculate single colonies of Agrobacterium containing pTRV1 and pTRV2-derived vectors in 5 mL YEP medium with appropriate antibiotics (kanamycin 50 μg/mL, rifampicin 25 μg/mL)
- Grow cultures at 28°C with shaking at 200 rpm for 24 hours
- Subculture 1 mL of the starter culture into 50 mL of fresh YEP medium with antibiotics and grow until OD600 reaches 0.6-0.8
- Pellet cells by centrifugation at 3,000 × g for 15 minutes and resuspend in MMA infiltration buffer (10 mM MES, 10 mM MgCl2, 200 μM acetosyringone, pH 5.6) to a final OD600 of 0.3-0.5 [1]
- Incubate the resuspended Agrobacterium cultures at room temperature for 3-4 hours without shaking
Plant Material Preparation and Agroinfiltration
Soybean Seed Sterilization and Germination:
- Surface-sterilize soybean seeds (cultivar Tianlong 1 has shown 95% efficiency) [1] in 70% ethanol for 2 minutes, followed by 2% sodium hypochlorite for 10 minutes with gentle agitation
- Rinse seeds thoroughly 4-5 times with sterile distilled water
- Soak sterilized seeds in sterile water for 12-16 hours until fully imbibed
- Aseptically bisect the swollen seeds longitudinally to obtain half-seed explants, ensuring the cotyledonary node remains intact on both halves
Agroinfiltration via Cotyledon Node:
- Combine equal volumes of Agrobacterium suspensions containing pTRV1 and pTRV2-derived vectors (e.g., pTRV2-GmPDS for photobleaching control)
- Immerse the prepared half-seed explants in the mixed Agrobacterium suspension for 20-30 minutes with occasional gentle agitation [1]
- Blot-dry the infected explants on sterile filter paper and transfer to co-cultivation medium (MS basal medium with 200 μM acetosyringone)
- Incubate in the dark at 25°C for 2-3 days to allow T-DNA transfer and initial infection
Plant Growth and Silencing Validation
Transplant and Growth Conditions:
- Transfer the co-cultivated explants to sterile soil mixture in growth chambers maintained at 22-24°C with 16-hour light/8-hour dark photoperiod
- Maintain high humidity (70-80%) for the first week after transplantation
- Monitor plants daily for development of silencing phenotypes, which typically appear 14-21 days post-infiltration (dpi) [1]
Efficiency Assessment Methods:
- For visible markers like GmPDS silencing, monitor photobleaching symptoms beginning at cluster buds and expanding to new leaves [1]
- Verify silencing efficiency at molecular level using quantitative PCR (qPCR) to measure transcript abundance of target genes
- For non-visual targets, include positive control plants infected with TRV2-GmPDS to confirm system functionality
- Calculate silencing efficiency as percentage of plants showing expected phenotype or significant transcript reduction (typically 65-95% with this protocol) [1]
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagents for Cotyledon Node VIGS in Soybean
| Reagent/Vector | Function/Purpose | Key Characteristics | Application Notes |
|---|---|---|---|
| pTRV1 & pTRV2 Vectors | TRV-based silencing system | Bipartite RNA virus; TRV1 encodes replication proteins, TRV2 carries target gene insert [7] | Required in 1:1 ratio for effective silencing |
| Agrobacterium tumefaciens GV3101 | Plant transformation vehicle | Disarmed strain with modified Ti plasmid; high transformation efficiency [1] [11] | Optimal OD600 0.3-0.5 for soybean cotyledon infiltration |
| MMA Infiltration Buffer | Agrobacterium resuspension medium | Contains MgCl₂, MES, and acetosyringone [1] | Acetosyringone induces vir genes essential for T-DNA transfer |
| Phytoene Desaturase (GmPDS) | Visual marker for silencing | Silencing causes photobleaching (white tissues) [1] | Critical positive control for protocol validation |
| MS Basal Medium | Co-cultivation substrate | Provides essential nutrients for plant cells [1] | Supplement with acetosyringone during co-cultivation phase |
Molecular Mechanism of VIGS
The efficacy of VIGS stems from its exploitation of the plant's conserved RNAi machinery. The molecular pathway can be visualized as a sequential process that initiates with vector delivery and culminates in targeted gene silencing.
Diagram 2: Molecular Pathway of VIGS. This diagram illustrates the sequential molecular events from viral vector delivery to phenotypic manifestation of gene silencing, highlighting the hijacking of the plant's native RNAi machinery.
The process begins with the delivery of TRV vectors containing target gene fragments to plant cells via Agrobacterium-mediated transformation. Following viral replication, double-stranded RNA forms as a replication intermediate, which is recognized by plant Dicer-like enzymes. These ribonucleases process the dsRNA into 21-24 nucleotide small interfering RNAs (siRNAs), which are then incorporated into the RNA-induced silencing complex (RISC). The guide strand of the siRNA directs RISC to complementary endogenous mRNA transcripts, resulting in sequence-specific cleavage and subsequent degradation. This targeted degradation depletes functional mRNA, leading to reduced protein levels and the manifestation of silencing phenotypes [7].
Applications in Soybean Research
The cotyledon node VIGS delivery method has enabled rapid functional characterization of genes involved in various biological processes in soybean:
Disease Resistance Gene Validation
VIGS has proven particularly valuable for studying soybean-pathogen interactions. Researchers have successfully silenced the rust resistance gene GmRpp6907, resulting in compromised resistance to Asian soybean rust [1]. Similarly, silencing of GmRPT4, a defense-related gene, confirmed its role in disease resistance mechanisms [1]. In brown stem rot resistance studies, VIGS was instrumental in demonstrating the oligogenic inheritance of resistance, revealing that at least two genes confer Rbs1-mediated resistance to Phialophora gregata [10].
Nodulation and Symbiosis Studies
The application of VIGS has extended to soybean-microbe interactions, particularly nodulation. A recently developed cowpea severe mosaic virus-based VIGS protocol has enabled functional analysis of genes involved in autoregulation of nodulation (AON), a process controlling optimal nodule number through systemic root-shoot-root signaling [9]. This represents a significant advancement as many AON pathway components expressed in aerial plant tissues remained understudied due to limitations of traditional root transformation approaches.
Specialized Metabolism and Stress Responses
Beyond disease resistance, cotyledon-based VIGS has been successfully applied to study specialized metabolic pathways in various plants, including medicinal species [11]. This demonstrates the methodology's versatility for investigating diverse biological processes, suggesting potential applications for studying soybean metabolic pathways involved in oil and protein biosynthesis, as well as abiotic stress responses.
Troubleshooting and Optimization Guidelines
Successful implementation of cotyledon node VIGS requires attention to several critical parameters:
- Low Silencing Efficiency: Optimize Agrobacterium density (OD600 0.3-0.5), extend immersion time to 30 minutes, and ensure explants contain intact cotyledonary nodes [1]
- Poor Viral Spread: Maintain high humidity (70-80%) during initial growth stages and verify plant vitality through proper nutrient and light conditions
- Inconsistent Phenotypes: Include positive controls (TRV2-GmPDS) in each experiment batch and use uniform plant developmental stages for infiltration
- High Mortality Rates: Reduce Agrobacterium density, minimize mechanical damage during explant preparation, and avoid prolonged immersion beyond 30 minutes [1]
The cotyledon node VIGS delivery method represents a robust platform for rapid gene function validation in soybean, significantly reducing the timeline for functional genomics studies compared to stable transformation. With silencing efficiencies reaching up to 95% in optimized conditions [1], this protocol provides researchers with a powerful tool to accelerate soybean genetic research and breeding programs.
Why the Cotyledon Node? Overcoming Barriers for Efficient Systemic Spread
Biological Rationale: The Cotyledon Node as a Strategic Gateway
The cotyledon node, also known as the cotyledonary node, is the region of a soybean seedling where the cotyledons (seed leaves) are attached to the embryonic axis. Its unique anatomical and physiological properties make it an exceptionally effective gateway for Virus-Induced Gene Silencing (VIGS) delivery, enabling robust systemic spread of the silencing signal throughout the plant.
The primary barrier to efficient VIGS in soybean is the plant's natural physical defenses, including a thick leaf cuticle and dense trichomes, which impede the penetration of Agrobacterium tumefaciens suspensions used in standard infiltration methods like spraying or injection [1]. The cotyledon node circumvents these barriers due to its distinct internal architecture. This region contains highly active meristematic cells and developing vascular tissues that are directly connected to the emerging primary shoot (plumule) and root (radicle) [8]. When the TRV vector is delivered via the cotyledon node, it gains direct access to this nascent vascular system. This access allows the virus to rapidly replicate and systemically traffic to newly developing tissues, including the first true leaves and the shoot apical meristem, achieving widespread gene silencing that is difficult to accomplish through other inoculation sites [1] [12].
The following diagram illustrates the logical relationship between the biological features of the cotyledon node and the resulting experimental advantages for VIGS.
Performance Data: Quantifying Silencing Efficiency
The establishment of a Tobacco Rattle Virus (TRV)-based VIGS system delivered via the cotyledon node has demonstrated high efficacy in silencing endogenous soybean genes. The silencing efficiency, confirmed by both phenotypic observation and molecular analysis, has been quantified for several key genes, as summarized in the table below [1].
Table 1: Silencing Efficiency of Endogenous Genes via Cotyledon Node-Based TRV-VIGS in Soybean
| Target Gene | Gene Function | Observed Phenotype | Time to Phenotype (Days Post-Inoculation) | Silencing Efficiency |
|---|---|---|---|---|
| GmPDS | Phytone desaturase (chlorophyll/carotenoid biosynthesis) | Photobleaching (white leaves) | 21 | 65% - 95% |
| GmRpp6907 | Rust resistance gene | Compromised rust immunity | Not Specified | 65% - 95% |
| GmRPT4 | Defense-related gene | Altered defense response | Not Specified | 65% - 95% |
The overall silencing efficiency for the system was reported to range from 65% to 95%, with an Agrobacterium infection efficiency that could reach up to 95% in the optimal soybean cultivar 'Tianlong 1' [1]. This high level of efficiency is a direct result of the effective systemic spread of the virus from the initial point of infection at the cotyledon node.
Experimental Protocol: Cotyledon Node-Based VIGS in Soybean
This section provides a detailed, step-by-step methodology for implementing the cotyledon node-based VIGS system in soybean, as established by recent research [1].
Required Materials and Reagents
Table 2: Research Reagent Solutions for Cotyledon Node-VIGS
| Item | Specification / Function | Details / Examples |
|---|---|---|
| VIGS Vectors | TRV-based binary vectors | pTRV1, pTRV2-GFP (with multiple cloning site) |
| Agrobacterium Strain | For plant transformation | Agrobacterium tumefaciens GV3101 |
| Soybean Genotype | Opt for high-efficiency cultivars | Tianlong 1, Williams 82 |
| Plant Growth Medium | Seed germination and explant culture | Murashige and Skoog (MS) basal medium |
| Antibiotics | Selection for bacterial and plant vectors | Kanamycin, Rifampicin |
| Induction Medium | Prepare Agrobacterium for infection | LB broth with antibiotics and MES buffer |
| Infiltration Solution | Resuspend bacteria for inoculation | MgCl₂, Acetosyringone |
Step-by-Step Workflow
The following diagram outlines the complete experimental workflow from vector construction to phenotypic analysis.
Part 1: Vector Construction and Agrobacterium Preparation
- Vector Construction: Clone a 200-400 bp fragment of the target soybean gene (e.g., GmPDS) into the multiple cloning site of the pTRV2 vector using appropriate restriction enzymes (e.g., EcoRI and XhoI) [1] [12]. The pTRV1 vector contains genes for viral replication and movement.
- Agrobacterium Transformation: Introduce the recombinant pTRV2 and the pTRV1 plasmids separately into Agrobacterium tumefaciens strain GV3101 using electroporation or freeze-thaw methods [1].
- Culture Preparation: Inoculate single colonies of Agrobacterium containing pTRV1 and pTRV2-target gene in separate liquid LB cultures with appropriate antibiotics. Grow overnight at 28°C with shaking.
- Induction: The next day, pellet the bacterial cultures by centrifugation and resuspend them in an induction buffer (e.g., 10 mM MgCl₂, 10 mM MES, pH 5.6, and 150 μM acetosyringone) to a final OD₆₀₀ of approximately 1.0. Incubate the suspensions at room temperature for 3-4 hours without shaking [1] [11].
- Inoculum Mixing: Combine the induced pTRV1 and pTRV2-target gene suspensions in a 1:1 ratio immediately before inoculation.
Part 2: Plant Inoculation and Analysis
- Plant Material Preparation: Surface-sterilize soybean seeds and soak them in sterile water for 24-48 hours in the dark until swollen. Under sterile conditions, longitudinally bisect the imbibed seeds to create half-seed explants, ensuring the cotyledon node remains intact on the explant [1] [13].
- Agroinfiltration: Immerse the fresh half-seed explants in the mixed Agrobacterium suspension for 20-30 minutes, ensuring full contact with the cotyledon node region [1].
- Co-cultivation and Growth: After infiltration, briefly blot the explants to remove excess liquid and transfer them to sterile filter paper or a semi-solid medium. Maintain the explants in a growth chamber with a 16/8-hour light/dark cycle at 22-25°C [1].
- Efficiency Validation:
- Infection Efficiency: Around 4 days post-infection (dpi), examine the cotyledon node under a fluorescence microscope for GFP signals to confirm successful Agrobacterium infection [1].
- Silencing Phenotype: Monitor plants for the appearance of visual silencing phenotypes (e.g., photobleaching for GmPDS) from 14 to 21 dpi [1].
- Molecular Confirmation: Use quantitative real-time PCR (qRT-PCR) on tissue samples from silenced leaves to quantify the reduction in endogenous target gene mRNA levels compared to control plants inoculated with an empty TRV vector [1] [12].
The cotyledon node is a critical tissue that overcomes the major physical and physiological barriers to efficient VIGS in soybean. Its meristematic nature and integration into the plant's developing vascular system provide a direct conduit for Agrobacterium delivery and the subsequent systemic spread of the TRV vector. The optimized protocol outlined here, leveraging this biological gateway, enables researchers to achieve high-efficiency gene silencing, thereby accelerating functional genomics and the discovery of agronomically important traits in soybean.
Virus-induced gene silencing (VIGS) has emerged as a powerful reverse genetics tool for rapid functional analysis of genes in plants, particularly in species like soybean where stable genetic transformation remains time-consuming and technically challenging [1] [14]. This technology exploits the plant's natural RNA interference (RNAi) machinery, using recombinant viral vectors to carry host gene fragments and trigger sequence-specific degradation of complementary mRNA targets [7] [15]. The application of VIGS in soybean research has accelerated the discovery of genes governing agronomically important traits, including disease resistance and stress tolerance [1] [16]. Among the various VIGS platforms developed, those based on Tobacco rattle virus (TRV), Bean pod mottle virus (BPMV), Apple latent spherical virus (ALSV), and Soybean yellow common mosaic virus (SYCMV) have shown significant utility in functional genomics studies [1] [14] [16]. This article provides a comprehensive comparative analysis of these four prominent VIGS vectors, with particular emphasis on their application in soybean research using cotyledon node delivery systems.
Comparative Analysis of Major VIGS Vectors
Table 1: Comparative characteristics of major VIGS vectors used in soybean functional genomics
| Vector | Virus Type | Delivery Methods | Silencing Efficiency | Key Advantages | Major Limitations |
|---|---|---|---|---|---|
| TRV | Positive-sense RNA virus (bipartite) | Agrobacterium-mediated cotyledon node immersion [1] | 65-95% [1] | Mild symptoms, broad tissue range including meristems, high efficiency [1] [15] | Limited application history in soybean [1] |
| BPMV | Positive-sense RNA virus | Particle bombardment [14] | Not explicitly quantified in sources | Well-established, widely adopted in soybean community [1] [16] | Technical hurdles, leaf phenotypic alterations [1] |
| ALSV | - | - | - | Effective in select varieties [14] | Narrow host range, limited to few soybean accessions [14] |
| SYCMV | Positive-sense RNA virus | - | - | Soybean-specific virus [16] | Less extensively documented in literature |
| CPSMV | Positive-sense RNA virus (bipartite) | Agro-infiltration of N. benthamiana followed by mechanical inoculation of soybean [14] | Robust silencing demonstrated [14] | Convenient propagation via N. benthamiana, versatile for VIGS and protein expression [14] | Relatively new vector, requires validation across diverse soybean genotypes |
Table 2: Molecular characteristics and experimental parameters of TRV and BPMV VIGS vectors
| Parameter | TRV-Based System | BPMV-Based System |
|---|---|---|
| Vector Construction | Two separate vectors: TRV1 (replicase components) and TRV2 (capsid protein + target insert) [1] | - |
| Insert Size | 300-500 bp fragments used [1] | - |
| Target Genes Validated | GmPDS, GmRpp6907 (rust resistance), GmRPT4 (defense-related) [1] | GmFNR (ferredoxin-NADP reductase) [16] |
| Optimal Agroinfiltration OD600 | 1.5 [17] | - |
| Time to Phenotype | Photobleaching observed at 21 dpi [1] | - |
| Key Applications | Disease resistance gene discovery, functional validation of candidate genes [1] | SMV resistance studies, photosynthesis-related gene function [16] |
The TRV-VIGS Protocol for Soybean via Cotyledon Node Delivery
Principle and Workflow
The TRV-VIGS system utilizes a bipartite vector system consisting of TRV1, encoding replication and movement proteins, and TRV2, containing the coat protein and a multiple cloning site for insertion of target gene fragments [15]. When delivered into plant cells via Agrobacterium tumefaciens, these vectors initiate viral replication and systemic movement, triggering the plant's RNAi machinery and resulting in degradation of mRNAs homologous to the inserted target sequence [15].
Step-by-Step Protocol
Vector Construction andAgrobacteriumPreparation
- Target Fragment Amplification: Amplify a 300-500 bp fragment of the target gene (e.g., GmPDS) using gene-specific primers with appropriate restriction sites (e.g., EcoRI and XhoI) [1].
- Cloning into TRV2 Vector: Ligate the purified PCR product into the corresponding sites of the pTRV2-GFP vector [1].
- Transformation: Introduce the recombinant plasmid into Agrobacterium tumefaciens strain GV3101 through standard transformation procedures [1].
- Culture Preparation: Grow positive Agrobacterium clones in YEB medium containing appropriate antibiotics (kanamycin 25 μg/mL and rifampicin 50 μg/mL) at 28°C with shaking until OD600 reaches 0.9-1.0 [18].
Plant Material Preparation and Inoculation
- Seed Sterilization and Germination: Surface-sterilize soybean seeds and germinate under sterile conditions until cotyledons are fully expanded [1] [17].
- Agroinfiltration Mixture Preparation: Mix Agrobacterium cultures containing pTRV1 and recombinant pTRV2 in a 1:1 ratio, resuspend in infiltration medium (10 mM MgCl2, 10 mM MES, 200 μM acetosyringone) to final OD600 of 1.5, and incubate for 3-4 hours at room temperature [1] [17].
- Cotyledon Node Inoculation:
- For soybean, bisect sterilized seeds longitudinally to obtain half-seed explants [1].
- Immerse fresh explants in the Agrobacterium suspension for 20-30 minutes, ensuring complete contact with the cotyledon node region [1].
- Alternatively, inject the Agrobacterium mixture directly into the cotyledon node using a needleless syringe [17].
Post-Inoculation Procedures and Analysis
- Plant Maintenance: Transfer inoculated plants to a growth chamber maintained at 23°C with a 16-hour light/8-hour dark photoperiod [17].
- Efficiency Assessment: At 4 days post-inoculation (dpi), examine infection efficiency by detecting GFP fluorescence under a microscope in hypocotyl sections [1].
- Phenotypic Monitoring: Observe systemic silencing phenotypes (e.g., photobleaching for GmPDS) beginning at 15-21 dpi [1] [17].
- Molecular Validation: Quantify silencing efficiency by qRT-PCR analysis of target gene expression in newly emerged leaves, comparing to control plants inoculated with empty TRV vectors [1].
Strategic Considerations for Vector Selection
The Scientist's Toolkit: Essential Reagents for TRV-VIGS in Soybean
Table 3: Essential research reagents for TRV-VIGS implementation in soybean
| Reagent/Resource | Function/Purpose | Specifications/Alternatives |
|---|---|---|
| pTRV1 and pTRV2 Vectors | Bipartite TRV vector system; TRV1 encodes replication proteins, TRV2 contains cloning site for target gene [1] | Available with GFP markers for tracking (pTRV2-GFP) [1] |
| Agrobacterium tumefaciens GV3101 | Delivery vehicle for introducing TRV vectors into plant cells [1] | Other strains (e.g., LBA4404) may require optimization |
| Soybean Cultivar Tianlong 1 | Optimized for TRV-VIGS with high infection efficiency (up to 95%) [1] | Other cultivars may require protocol adjustment |
| Infiltration Medium | Resuspension medium for Agrobacterium during inoculation | 10 mM MgCl₂, 10 mM MES, 200 μM acetosyringone, pH 5.6 [1] |
| Marker Genes (GmPDS, GmCLA1) | Positive controls for optimizing silencing efficiency; produce visible photobleaching phenotype [1] [17] | ZjPDS used in jujube, NbPDS in N. benthamiana [17] |
| Restriction Enzymes (EcoRI, XhoI) | Insertion of target gene fragments into TRV2 vector [1] | Gateway cloning alternatives available [15] |
The comparative analysis of VIGS vectors presented herein highlights the distinct advantages and limitations of TRV, BPMV, ALSV, and SYCMV systems for soybean functional genomics. The TRV-based system emerges as particularly promising due to its high silencing efficiency, mild symptomatic effects, and adaptability to Agrobacterium-mediated cotyledon node delivery. The optimized protocol detailed in this article provides researchers with a robust framework for implementing TRV-VIGS in soybean studies, potentially accelerating the discovery and validation of genes controlling agronomically important traits. As VIGS technology continues to evolve, integration with emerging genome-editing platforms and multi-omics approaches will further enhance its utility in soybean improvement programs.
A Step-by-Step Protocol for TRV-Mediated VIGS via Cotyledon Node Delivery
Within the framework of cotyledon node-based Virus-Induced Gene Silencing (VIGS) delivery in soybean research, the construction of recombinant viral vectors represents a critical initial step. The pTRV2-GFP vector serves as a versatile backbone for inserting target gene fragments, enabling both efficient gene silencing and visual tracking of viral spread through Green Fluorescent Protein (GFP) expression [1] [15]. This system leverages the Tobacco Rattle Virus (TRV)-based VIGS mechanism, which triggers post-transcriptional gene silencing by processing double-stranded RNA into siRNAs that guide the degradation of complementary endogenous mRNA sequences [19] [7] [15]. The following application note details a standardized protocol for cloning target fragments into the pTRV2-GFP vector, optimized specifically for soybean functional genomics studies using cotyledon node delivery.
Research Reagent Solutions
Table 1: Essential reagents and materials for pTRV2-GFP vector construction and VIGS in soybean.
| Reagent/Material | Specification/Function |
|---|---|
| pTRV2-GFP Vector | Binary VIGS vector containing GFP reporter for tracking viral infection [1]. |
| Restriction Enzymes | EcoRI and XhoI for directional cloning of target fragments into the MCS [1]. |
| Agrobacterium tumefaciens | Strain GV3101 for plant transformation; delivers T-DNA containing VIGS constructs [1] [20]. |
| Plant Gene Fragment | 300-500 bp target gene sequence (e.g., GmPDS, GmRpp6907) amplified from soybean cDNA [1]. |
| Gateway Cloning System | Alternative system using attB/attR site-specific recombination (e.g., BP Clonase enzyme) [15]. |
Protocol: Cloning Target Fragments into the pTRV2-GFP Vector
Fragment Amplification and Vector Preparation
The initial phase involves the isolation of the target gene fragment and preparation of the vector backbone for ligation.
- Primer Design and Fragment Amplification: Design gene-specific primers to amplify a 300-500 base pair fragment from the target gene (e.g., GmPDS). The primers must incorporate appropriate restriction enzyme sites at their 5' ends for subsequent directional cloning.
- Vector Digestion: Digest the pTRV2-GFP plasmid with the EcoRI and XhoI restriction enzymes. Purify the linearized vector fragment using a standard gel extraction kit to prevent self-ligation [1].
- Ligation and Transformation: Ligate the purified PCR product into the prepared pTRV2-GFP backbone using T4 DNA ligase. Transform the ligation product into competent E. coli cells (e.g., DH5α) and select positive clones on LB agar plates containing kanamycin (50 mg/L) [1].
- Sequence Verification: Isolate plasmid DNA from positive colonies and verify the correct insertion and sequence of the target fragment via Sanger sequencing [1].
Agrobacterium Transformation and Cotyledon Node VIGS in Soybean
Following vector construction, the recombinant plasmid is introduced into Agrobacterium for plant delivery.
- Agrobacterium Transformation: Introduce the verified recombinant pTRV2-GFP plasmid and the helper plasmid pTRV1 into Agrobacterium tumefaciens strain GV3101 via electroporation or the freeze-thaw method [1] [20].
- Soybean Cotyledon Node Infection:
- Prepare Agrobacterium cultures (harboring both pTRV1 and the recombinant pTRV2-GFP) by growing them to an optimal optical density of OD₆₀₀ = 1.5 [19] [1].
- Bisect sterilized soybean seeds longitudinally to create half-seed explants containing the cotyledon node.
- Immerse the fresh explants in the Agrobacterium suspension for 20-30 minutes to facilitate infection [1].
- Efficiency Evaluation and Plant Growth: Post-infection, excise a portion of the hypocotyl and observe under a fluorescence microscope to detect GFP signals, confirming successful infection. Transplant treated seedlings to soil and maintain under controlled environmental conditions. Silencing phenotypes, such as photobleaching from GmPDS knockdown, typically become visible within 21 days post-inoculation (dpi) [1].
Key Experimental Data and Parameters
Table 2: Key quantitative parameters for efficient TRV-VIGS in soybean via cotyledon node delivery.
| Parameter | Optimal Condition/Specification | Experimental Context |
|---|---|---|
| Target Fragment Length | 193–500 bp [1] [20] | A 193-bp HaPDS fragment was effective in sunflower [20]. |
| Agrobacterium OD₆₀₀ | 1.5 [19] [1] | Higher than the OD=1.0 used for N. benthamiana [19]. |
| Infection Duration | 20–30 min (immersion) [1] | For cotyledon node explants. |
| Silencing Onset | ~21 days post-inoculation (dpi) [1] | Phenotypic observation (e.g., photobleaching). |
| Silencing Efficiency | 65%–95% [1] | Range observed in soybean cultivar 'Tianlong 1'. |
| Transformation Efficiency | >80% (up to 95%) [1] | Based on GFP fluorescence observation in cultivar 'Tianlong 1'. |
Workflow and Mechanism Visualization
Diagram 1: Experimental workflow for constructing the pTRV2-GFP VIGS vector and its subsequent application in soybean. The process begins with molecular cloning to create the recombinant vector, followed by its delivery into soybean plants via Agrobacterium to initiate the cellular gene silencing mechanism.
Within the broader scope of establishing a virus-induced gene silencing (VIGS) system in soybean via cotyledon node delivery, the preparation of Agrobacterium tumefaciens is a critical foundational step. The selection of an appropriate bacterial strain and the optimization of its culture conditions directly determine the efficiency of T-DNA transfer into plant cells, which is paramount for achieving high silencing efficiency in subsequent functional genomics studies. This protocol details evidence-based methods for strain selection and culture preparation, specifically contextualized for soybean VIGS research.
Key Research Reagent Solutions
The table below catalogues essential reagents and their specific functions in preparing Agrobacterium for soybean transformation.
Table 1: Essential Research Reagents for Agrobacterium Preparation
| Reagent | Function/Application | Key Details |
|---|---|---|
| Agrobacterium Strains | Delivery of T-DNA containing VIGS constructs into plant cells. | GV3101, AGL1, and EHA105 are common hypervirulent strains [1] [21] [22]. |
| Antibiotics | Selection of transformed Agrobacterium and control of bacterial growth post-co-cultivation. | Strain-dependent (e.g., Kanamycin, Carbenicillin, Rifampicin, Gentamycin) [1] [23]. |
| Acetosyringone | Phenolic inducer of Agrobacterium Vir genes, enhancing T-DNA transfer competence. | Typically used at 100-200 µM in induction and co-cultivation media [21] [24] [25]. |
| AB Minimal Salts | Component of the induction medium for enhancing bacterial virulence. | Used in AB-MES induction medium [21]. |
| Pluronic F68 | Surfactant that can enhance transformation efficiency. | Added to co-cultivation medium at 0.05% (w/v) [21]. |
Strain Selection and Culture Media
The choice of Agrobacterium strain is a primary determinant of transformation success. Different strains exhibit varying levels of virulence and host compatibility.
Table 2: Agrobacterium Strain Selection for Plant Transformation
| Strain | Key Characteristics | Documented Use in Literature |
|---|---|---|
| GV3101 | A disarmed strain known for high efficiency in transient transformation and VIGS. | Utilized in TRV-based VIGS in soybean and tobacco ringspot virus (TRSV)-based VIGS [1] [25]. |
| AGL1 | Hypervirulent strain carrying a C58 chromosome background; often provides high transformation efficiency, including in recalcitrant species. | Achieved near 100% transient transformation in Arabidopsis suspension cells and efficient stable transformation in grapevine and poplar [21] [24] [22]. |
| EHA105 | A hypervirulent derivative of the EHA101 strain, widely used for stable transformation in diverse crops. | Successfully used for stable transformation in Jonquil and grapevine [26] [22]. |
Culture Media Composition
- Standard Growth Medium: For routine growth and plasmid maintenance, Luria-Bertani (LB) or YEB medium is used, supplemented with the appropriate antibiotics [21] [23].
- Induction Medium: To activate the bacterial virulence system, bacteria are resuspended in an induction medium prior to plant inoculation. A common and effective formulation is AB-MES medium [21].
- AB-MES Medium: 50% (v/v) AB minimal salts, 1.1 g/L MS basal salts, 0.25% (w/v) sucrose, pH 5.5 [21]. The acidic pH and the presence of acetosyringone are critical for inducing the Vir genes.
Quantitative Experimental Data
Optimizing the physical parameters of the bacterial culture is essential for maximizing transformation efficiency while minimizing detrimental density-dependent effects.
Table 3: Optimized Agrobacterium Culture Conditions
| Parameter | Optimal Range | Experimental Context & Impact |
|---|---|---|
| Optical Density (OD600) | 0.3 - 0.8 | An OD600 of 0.6 was optimal for poplar callus transformation [24]. Higher densities can lead to antagonism, reducing per-bacterium transformation efficiency [27]. |
| Acetosyringone Concentration | 100 - 200 µM | 100 µM was used for poplar transformation [24], while 200 µM was effective for Arabidopsis and Nicotiana benthamiana transformation [21] [23]. |
| Co-cultivation Duration | 2 - 3 days | A 2-day co-cultivation period was standard for several protocols, including poplar and N. benthamiana [21] [24]. |
Step-by-Step Protocol for Agrobacterium Preparation
This protocol outlines the preparation of an Agrobacterium culture suitable for infecting soybean cotyledon nodes in a VIGS assay.
Materials
- Agrobacterium tumefaciens strain (e.g., GV3101) harboring the VIGS binary vector (e.g., pTRV1, pTRV2-GOI) [1].
- LB solid and liquid media with appropriate antibiotics (e.g., 50 µg/mL Kanamycin, 50 µg/mL Rifampicin).
- Antibiotic stock solutions.
- Induction medium (e.g., ABM-MS or MgCl2/MES buffer).
- 200 mM acetosyringone stock solution (in DMSO or ethanol).
- Sterile centrifuge tubes.
- Benchtop centrifuge.
- Spectrophotometer.
Workflow
Detailed Methodology
Strain Revival and Preculture
- Streak the Agrobacterium strain from a -80°C glycerol stock onto a fresh LB agar plate containing the requisite antibiotics. Incubate the plate at 28°C for 48-72 hours until single colonies form [21].
- Pick a single, well-isolated colony and inoculate it into a culture tube containing 3-5 mL of liquid LB medium with the same antibiotics. Incubate the tube overnight (16-20 hours) at 28°C with constant shaking at 160-200 rpm [23].
Main Culture and Virulence Induction
- The following day, measure the OD600 of the preculture. Use this to inoculate a larger volume of induction medium (e.g., ABM-MS) to a starting OD600 of approximately 0.2. Supplement the induction medium with 200 µM acetosyringone to activate the bacterial virulence genes [21].
- Incubate the main culture again at 28°C with shaking until the OD600 reaches the target range of 0.3 to 0.8. This typically takes 4-6 hours.
Harvesting and Preparation for Inoculation
- Transfer the bacterial culture to sterile centrifuge tubes and pellet the cells by centrifugation at 6,800 × g for 10 minutes at room temperature [21].
- Carefully decant the supernatant and resuspend the bacterial pellet in the desired infiltration buffer or fresh induction medium. A common buffer is 10 mM MgCl2 with 10 mM MES, pH 5.5-5.7, supplemented with 150-200 µM acetosyringone [23] [25].
- Adjust the final OD600 of the suspension to the required density for your specific inoculation method. For cotyledon node immersion, a final OD600 of 0.8 has been successfully employed [1].
Troubleshooting and Technical Notes
- Bacterial Density Antagonism: At high total bacterial densities (OD600 > 1.0), bacteria exhibit antagonistic interactions that can significantly reduce the transformation efficiency per bacterium [27]. It is crucial to use the lowest effective OD for inoculation.
- Culture Purity: Always start from a single colony to ensure a genetically uniform culture and prevent overgrowth by non-transformed cells.
- Acetosyringone Stability: Acetosyringone is light-sensitive and can degrade. Prepare stock solutions fresh or store aliquots at -20°C protected from light.
- Plant Material Compatibility: The thick cuticle and dense trichomes on soybean leaves can impede liquid penetration. Therefore, for cotyledon node delivery, the immersion method is more effective than leaf infiltration or misting [1].
Within the broader scope of a thesis investigating cotyledon node VIGS delivery in soybean, the preparation of high-quality, sterile plant material is a critical first step. The half-seed explant method, which exposes the cotyledonary node, is a established technique for efficient Agrobacterium-mediated transformation and virus delivery [1] [28]. This protocol outlines a standardized procedure for soybean seed sterilization and the generation of half-seed explants, optimized for subsequent VIGS experiments.
Reagent and Equipment Setup
Research Reagent Solutions
Table 1: Essential reagents and materials for seed sterilization and explant preparation.
| Item Name | Function / Purpose | Technical Specification / Notes |
|---|---|---|
| Soybean Seeds | Plant material for generating explants. | Ensure seeds are from a sterilized soil source to minimize endogenous contamination [28]. |
| Commercial Bleach | Primary surface disinfectant. | A 20-30% solution is typically used [28] [29]. |
| 12N HCl | Generates chlorine gas for sterilization. | Added slowly to bleach in a fume hood for gas sterilization [28]. Caution: Corrosive. |
| Sterile Deionized Water | Rinsing seeds to remove residual sterilants. | Must be autoclaved to maintain sterility [28] [30]. |
| 0.9% Saline Solution | Initial wash to remove debris from nodules or seeds. | Autoclaved before use [30]. |
| Gamborg's B5 Medium | Basal medium for subsequent tissue culture steps. | Provides essential nutrients for explant co-cultivation and growth [28]. |
| L-Cysteine & Sodium Thiosulfate | Anti-browning agents in co-cultivation media. | Prevents tissue necrosis by mitigating oxidative stress during Agrobacterium co-cultivation [28]. |
Step-by-Step Protocol
Seed Sterilization
This procedure utilizes chlorine gas, an effective method for sterilizing seeds with complex surface textures [28].
- Preparation: Place dry soybean seeds in an open Petri dish within a glass desiccator.
- Gas Generation: In a fume hood, add ~100 mL of commercial bleach to a 150 mL beaker inside the desiccator. Slowly add 3.5 mL of 12N HCl to the bleach. Immediately seal the desiccator.
- Sterilization: Leave the sealed desiccator overnight at room temperature.
- Aeration: The next day, transfer the Petri dish to a laminar flow hood. Remove the lid and let the seeds air out for 30 minutes to dissipate residual chlorine gas [28].
- Storage: Seal the plate with micropore tape and store at room temperature until needed.
Half-Seed Explant Preparation
The objective is to obtain a clean explant with the cotyledonary node exposed, ready for Agrobacterium infection in VIGS protocols [1] [28].
- Imbibition: In a laminar flow hood, add sterile deionized water to the surface-sterilized seeds until submerged. Cover the plate with foil to protect from light and incubate at room temperature for 20 hours [28].
- Dissection:
- Transfer an imbibed seed to a sterile Petri dish.
- Using a sterile scalpel, make a longitudinal cut along the hilum to separate the two cotyledons [28].
- Carefully remove the seed coat.
- Excise the embryonic axis (the radicle and plumule) found at the nodal end of the cotyledons.
- Inspect the cotyledonary node and remove any remaining axial buds attached to it [28].
- Resulting Explant: The final product is a "half-seed" with a flat, exposed cotyledonary node region. Multiple explants should be placed in a sterile dish for the subsequent infection step.
Diagram 1: Half-seed explant generation workflow.
Application in VIGS Research
The half-seed explant is particularly suited for VIGS studies due to its high regeneration potential and accessibility for Agrobacterium delivery. The cotyledonary node contains actively dividing cells that are ideal for virus replication and systemic spread of the silencing signal [1] [8].
Table 2: Key metrics for TRV-VIGS delivery in soybean half-seed explants.
| Parameter | Quantitative Data / Observation | Significance for VIGS |
|---|---|---|
| Agrobacterium Infection Efficiency | >80% of cells show fluorescence signal post-infection [1]. | High infection rate is crucial for successful VIGS vector delivery. |
| Silencing Onset | Phenotypes (e.g., photobleaching) observed at 21 days post-inoculation (dpi) [1]. | Informs the experimental timeline for phenotypic analysis. |
| Systemic Silencing Efficiency | Ranges from 65% to 95% for endogenous genes like GmPDS [1]. | Demonstrates the robustness of the system for functional genomics. |
Diagram 2: Explant use in TRV-VIGS workflow.
Within the broader scope of cotyledon node Virus-Induced Gene Silencing (VIGS) delivery in soybean research, the Agrobacterium immersion method stands out as a highly efficient technique for systemic gene silencing. This protocol addresses a significant challenge in soybean functional genomics: the difficulty of achieving efficient gene silencing due to the plant's thick cuticle and dense leaf trichomes, which impede liquid penetration using conventional methods like misting or direct injection [1]. The core innovation of this approach involves using bisected cotyledonary explants immersed in an Agrobacterium tumefaciens suspension containing tobacco rattle virus (TRV) vectors, enabling robust infection and systemic spread of the silencing effect throughout the plant [1]. This method establishes a critical platform for rapid functional characterization of genes involved in disease resistance and stress tolerance in soybean.
Key Reagents and Equipment
The successful implementation of the cotyledon node immersion protocol requires the following essential research reagent solutions and laboratory equipment.
Table 1: Essential Research Reagent Solutions
| Item Name | Function/Description |
|---|---|
| Agrobacterium tumefaciens GV3101 | Bacterial strain used as the delivery vehicle for the TRV vectors [1] [31]. |
| pTRV1 and pTRV2 Vectors | Bipartite tobacco rattle virus (TRV) system. TRV1 contains replication proteins, while TRV2 carries the coat protein and the inserted target gene fragment for silencing [1] [31]. |
| Luria-Bertani (LB) Medium | Standard microbial growth medium for cultivating Agrobacterium strains [31]. |
| Antibiotics (Kanamycin, Gentamycin) | Added to the LB medium to maintain selective pressure and ensure the retention of the TRV vectors in the Agrobacterium culture [31]. |
| Acetosyringone | A phenolic compound that induces the Agrobacterium Vir genes, enhancing the efficiency of T-DNA transfer into the plant cells [1]. |
| Murashige and Skoog (MS) Medium | A basal plant culture medium used for seed germination and/or maintaining explants post-infection [1]. |
Table 2: Necessary Laboratory Equipment
| Item Name | Function/Description |
|---|---|
| Sterile Tissue Culture Supplies | Including Petri dishes, containers, and tools for aseptic handling of plant explants. |
| Fluorescence Microscope | Essential for evaluating initial infection efficiency by detecting GFP fluorescence at the cotyledonary node 4 days post-infection [1]. |
| Vacuum Infiltration Apparatus (Optional) | While the standard immersion method is effective, some protocols for other plant species use vacuum infiltration to improve Agrobacterium penetration, especially in intact tissues [11]. |
Experimental Protocol
Agrobacterium Culture Preparation
- Transformation: Introduce the pTRV1 and pTRV2-derived plasmids (e.g., pTRV2–GFP, pTRV2–GFP-GmPDS) into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method [31].
- Starter Culture: Inoculate a single colony of transformed Agrobacterium into a small volume (e.g., 1-2 mL) of LB liquid medium containing the appropriate antibiotics (e.g., Kanamycin and Gentamycin, both at 50 mg/L). Incubate overnight at 28°C with constant shaking at 200-220 rpm [1] [31].
- Secondary Culture: Dilute the overnight culture (e.g., 200 µL) into a larger volume of fresh, antibiotic-supplemented LB medium (e.g., 10-50 mL). Continue incubation until the culture reaches the optimal density for infection.
- Induction and Preparation: Pellet the bacterial cells by centrifugation (e.g., 3000-5000 rpm for 10-15 minutes). Resuspend the pellet in an induction buffer (e.g., 10 mM MgCl₂, 10 mM MES, pH 5.6) containing 150 µM acetosyringone. Adjust the final suspension to an optical density at 600 nm (OD600) of approximately 0.4-1.0 [1] [11]. Allow the induced suspension to incubate at room temperature for several hours (e.g., 2-4 hours) before use.
Plant Explant Preparation
- Seed Sterilization and Germination: Surface-sterilize soybean seeds (e.g., cultivar Tianlong 1) and allow them to soak in sterile water until they are fully swollen.
- Explant Generation: Under sterile conditions, longitudinally bisect the swollen seeds to obtain half-seed explants, each containing a cotyledon node [1]. These fresh, cut explants are ideal for infection.
Core Infection Process: Immersion
- Combining Agrobacterium and Explant: Mix the prepared Agrobacterium suspensions containing pTRV1 and pTRV2-derived vectors in a 1:1 ratio.
- Immersion: Submerge the freshly prepared half-seed explants completely in the mixed Agrobacterium suspension.
- Optimal Duration: Allow the immersion to proceed for 20 to 30 minutes [1]. This duration has been identified as optimal for achieving high infection efficiency without causing damage to the tissues.
- Co-cultivation: After immersion, briefly blot the explants to remove excess liquid and transfer them to a solid co-cultivation medium (e.g., containing MS salts and agar). Maintain the explants in the dark at room temperature for 2-3 days to facilitate T-DNA transfer.
Post-infection Procedures and Silencing Evaluation
- Plantlet Development: Following co-cultivation, transfer the explants to a tissue culture environment that promotes shoot and root development.
- Efficiency Check (at 4 dpi): To assess initial infection success, examine the cotyledonary nodes under a fluorescence microscope for GFP signals. A successful infection will show fluorescence in over 80% of cells in the transverse section [1].
- Phenotypic and Molecular Analysis (from 14-21 dpi): Monitor the developed plants for the emergence of silencing phenotypes, such as photobleaching in GmPDS-silenced plants, which typically becomes visible around 21 days post-inoculation (dpi) [1]. Confirm silencing efficiency through quantitative PCR (qPCR) to measure the reduction in target gene transcript levels, which can range from 65% to 95% with this method [1].
Figure 1: Experimental workflow for the cotyledon node immersion method, highlighting the core infection step and key evaluation timepoints.
Data Presentation and Analysis
The efficiency of the VIGS system is quantified through both phenotypic observation and molecular analysis. The data below summarizes key quantitative outcomes from implementing this protocol.
Table 3: Quantitative Data on Immersion-Based VIGS Efficiency
| Parameter Measured | Result / Value | Method of Assessment / Notes |
|---|---|---|
| Optimal Immersion Duration | 20 - 30 minutes | Determined as the timeframe yielding maximum infection efficiency without tissue damage [1]. |
| Initial Infection Efficiency | >80% of cells | Evaluated by GFP fluorescence in transverse sections of the cotyledon node at 4 days post-infection (dpi) [1]. |
| Overall Silencing Efficiency | 65% - 95% | Range of silencing efficiency measured by qPCR analysis of target gene transcripts in systemic leaves [1]. |
| Phenotype Onset | ~21 dpi | Time when visible silencing phenotypes (e.g., photobleaching) first appear systemically [1]. |
Figure 2: The molecular pathway of Virus-Induced Gene Silencing (VIGS) initiated by Agrobacterium delivery of TRV vectors, leading to systemic silencing.
Within the framework of a broader thesis investigating cotyledon node Virus-Induced Gene Silencing (VIGS) in soybean, the steps following Agrobacterium-mediated infection are critical for successful functional genomics research. The post-infection phase, encompassing tissue culture and the transition to soil, directly influences plant survival, silencing efficiency, and the robustness of subsequent phenotypic data. This protocol details a highly efficient TRV-based VIGS system for soybean, established through the infection of cotyledonary nodes, and provides a standardized workflow for researchers from infection to the acquisition of phenotype data [1].
Experimental Workflow and Key Observations
The entire process, from seed preparation to phenotypic analysis, is designed to be completed within approximately 30 days. The workflow below outlines the major stages and the typical timeline for key observations.
Detailed Protocol: From Tissue Culture to Soil
Tissue Culture Phase (Post-Infection)
Following agroinfiltration, the explants enter a tissue culture phase designed to recover and allow for the systemic spread of the virus and the establishment of silencing [1].
Step 1: Co-cultivation and Recovery
- After immersion in the Agrobacterium suspension, the infected half-seed explants should be blotted dry on sterile filter paper.
- Transfer the explants to a co-cultivation medium. The specific composition of this medium was not detailed in the search results, but standard soybean tissue culture protocols typically use a basal medium with minimal nutrients to support initial recovery without promoting excessive callus growth.
- Maintain the explants under sterile conditions in a growth chamber for 10-14 days. Recommended environmental conditions are a 16/8 hour light/dark photoperiod and a temperature of 25°C.
Step 2: Monitoring Infection Efficiency
- As a critical quality control step, infection efficiency can be evaluated non-destructively at 4 days post-infection (dpi) [1].
- Excise a small portion of the hypocotyl from select explants under a sterile microscope.
- Observe the tissue under a fluorescence microscope for the presence of GFP fluorescence. In the established protocol, more than 80% of cells in a transverse section typically show successful infection, confirming high efficiency [1].
Transitioning Plants to Soil
The transition from a sterile, high-humidity tissue culture environment to soil is a critical step to prevent transplant shock and ensure continued plant development for phenotypic assessment.
Step 1: Acclimatization
- Around 21-28 days post-inoculation (dpi), seedlings from the treatment group (e.g., pTRV:GmPDS) are ready for transplanting [1].
- Gently remove the plantlets from the culture medium, carefully rinsing any residual agar from the roots with sterile water to prevent microbial growth.
- Transfer the plantlets to small pots containing a pre-moistened, sterile nutrient soil mix. A mix suitable for soybean growth, such as one containing peat and perlite, is recommended.
Step 2: Post-Transplant Care
- To maintain high humidity initially, cover the pots with transparent plastic domes or clear plastic bags for the first 2-3 days.
- Gradually reduce humidity over the next 4-7 days by making small openings in the cover before removing it completely. This "hardening off" process allows the plants to adapt to ambient conditions.
- Grow the plants under controlled greenhouse or growth chamber conditions with a 16/8 hour light/dark cycle and a temperature of 25°C.
Phenotypic and Molecular Validation
Successful gene silencing is confirmed through a combination of visual phenotypes and molecular analysis.
Phenotypic Timeline and Silencing Efficiency
The table below summarizes the key milestones for observing and validating VIGS in soybean using the cotyledon node method.
Table 1: Key Observations and Silencing Efficiency in Soybean VIGS
| Days Post-Infection (dpi) | Key Observation/Milestone | Reported Efficiency/Outcome | Citation |
|---|---|---|---|
| 4 dpi | GFP fluorescence validation | >80% of cells show infection | [1] |
| 14 dpi | Initial silencing phenotype in cluster buds | Not specified (Early stage) | [1] |
| 21 dpi | Photobleaching in leaves (for GmPDS) | Clearly visible phenotype | [1] |
| Full Experiment | Systemic silencing efficiency | 65% to 95% | [1] |
Molecular Validation
- qPCR Analysis: To quantitatively confirm the downregulation of the target gene, conduct qPCR on leaf tissue samples collected from control (pTRV:empty) and silenced plants. A significant reduction in the transcript level of the target gene (e.g., GmPDS) is expected in successfully silenced plants [1].
- Phenotypic Scoring: For visible marker genes like GmPDS, the percentage of plants showing the characteristic photobleaching phenotype is a direct measure of silencing efficiency. The established protocol achieves a high efficiency range of 65% to 95% [1].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagent Solutions for Cotyledon Node VIGS in Soybean
| Reagent/Vector | Function/Description | Application in Protocol |
|---|---|---|
| Agrobacterium tumefaciens GV3101 | Bacterial strain used for vector delivery. | Harbors the TRV vectors; used to prepare the suspension for infecting explants. |
| pTRV1 and pTRV2 Vectors | Components of the bipartite Tobacco Rattle Virus (TRV) VIGS system. | pTRV2 is modified to carry the target gene fragment (e.g., GmPDS). Both are required for systemic infection and silencing. |
| Half-Seed Explants | Plant material containing the cotyledon node. | The specific explant type used for highly efficient Agrobacterium infection in this optimized protocol. |
| Sterile Tissue Culture Medium | Supports explant survival and recovery post-infection. | Used during the co-cultivation and recovery phase after agroinfiltration. |
| GFP Reporter Gene | Visual marker for assessing infection efficiency. | Cloned into the TRV vector; allows for fluorescence-based validation of successful infection at 4 dpi. |
Concluding Remarks
This application note provides a validated protocol for the post-infection handling of soybean plants subjected to cotyledon node-mediated VIGS. The key to success lies in the high-efficiency infection method using half-seed explants and the careful management of plants through the tissue culture and acclimatization phases. By adhering to this workflow, researchers can reliably achieve systemic gene silencing, enabling rapid functional characterization of soybean genes with implications for disease resistance and stress tolerance research [1].
In plant biotechnology research, particularly in studies involving Virus-Induced Gene Silencing (VICS) in soybean, confirming the success of the initial infection step is crucial for experimental validity. The use of Green Fluorescent Protein (GFP) as a visual reporter provides a rapid, non-destructive method to assess both the efficiency and the spatial pattern of infection before proceeding to more labor-intensive molecular analyses. Within the context of a broader thesis on cotyledon node VIGS delivery in soybean, this protocol details how to leverage GFP fluorescence for robust efficiency assessment. This method is especially valuable for optimizing delivery systems, such as Agrobacterium tumefaciens-mediated transformation of cotyledon nodes, where traditional confirmation methods are slow and destructive [1].
The fundamental principle relies on co-delivering a GFP gene construct alongside the VIGS vector. Successful Agrobacterium infection leads to GFP expression within plant cells, which can be visualized under specific light conditions. This allows researchers to quickly identify successfully treated explants, quantify infection rates, and select the best specimens for subsequent experiments, thereby saving time and resources [1] [32].
Key Concepts and Quantitative Benchmarks
Establishing a baseline for expected outcomes is key to interpreting GFP fluorescence results. The table below summarizes core efficiency metrics and challenges based on established protocols in soybean.
Table 1: Key Quantitative Metrics and Considerations for GFP-Based Infection Assessment
| Metric/Parameter | Typical Result/Value | Technical Notes |
|---|---|---|
| Infection Efficiency | 65% - 95% [1] | Efficiency is highly dependent on soybean cultivar, explant type, and Agrobacterium strain. |
| Time to Initial Detection | 2-4 days post-infection [1] | Fluorescence can often be detected within days, allowing for early screening. |
| Optimal Observation Site | Cotyledon node & hypocotyl [1] | The vascular-rich cotyledon node is the primary site for initial infection and GFP spread. |
| Spatial Pattern | Systemic spread from infection site [1] | In an optimized TRV-VIGS system, fluorescence spreads systemically from the cotyledon node. |
| Primary Challenge | GFP leakiness in unfixed tissue [32] | In snap-frozen or unfixed tissues, GFP can leak from cells, leading to diffuse signal and inaccurate localization. |
| Mitigation Strategy | Direct post-fixation with warm PFA [32] | Omitting a drying step and directly fixing cryosections with 4% PFA at 30-37°C preserves GFP localization. |
Experimental Protocols
Agrobacterium-Mediated Infection of Soybean Cotyledon Nodes
This protocol is adapted from a TRV-VIGS study in soybean, which achieved high infection efficiency [1].
Materials:
- Soybean seeds (e.g., cultivar 'Tianlong 1')
- Agrobacterium tumefaciens strain GV3101 harboring pTRV1 and pTRV2-GFP vectors
- Luria-Bertani (LB) broth and agar with appropriate antibiotics (e.g., kanamycin, rifampicin)
- Sterilization solution (e.g., 70% ethanol, commercial bleach)
- Co-cultivation medium (sterile, semi-solid)
Procedure:
- Seed Sterilization and Preparation: Surface-sterilize soybean seeds and soak in sterile water until swollen. Bisect the seeds longitudinally to obtain half-seed explants, ensuring the cotyledon node is intact [1].
- Agrobacterium Culture Preparation:
- Inoculate a single colony of Agrobacterium containing the pTRV2-GFP plasmid into LB broth with antibiotics.
- Incubate at 28°C with shaking (200 rpm) until the culture reaches an OD₆₀₀ of 0.4-0.8.
- Centrifuge the culture and resuspend the pellet in an induction medium (e.g., LB with 200 µM acetosyringone) to the final working OD₆₀₀ of ~0.5.
- Infection and Co-cultivation:
- Immerse the prepared half-seed explants in the Agrobacterium suspension for 20-30 minutes with gentle agitation [1].
- After infection, blot the explants dry on sterile filter paper and transfer them to co-cultivation medium.
- Incubate the plates in the dark at 22-25°C for 2-4 days.
GFP Fluorescence Evaluation and Imaging
This protocol covers the assessment of infection efficiency using fluorescence microscopy.
Materials:
- Fluorescence microscope equipped with a GFP filter set (excitation ~470 nm, emission ~525 nm)
- Sharp blade or microtome for sectioning
- Post-fixation solution: 4% Paraformaldehyde (PFA) in phosphate buffer, pre-warmed to 30-37°C [32]
Procedure:
- Sample Harvest: On the 4th day post-infection, excise a portion of the hypocotyl containing the cotyledon node under sterile conditions [1].
- Sectioning (Optional but Recommended):
- For precise cellular localization, prepare thin cross-sections or longitudinal sections of the cotyledon node region.
- To prevent GFP leakiness, omit any drying steps. Immediately immerse the freshly cut, unfixed sections into the pre-warmed 4% PFA solution for post-fixation [32].
- Microscopy and Image Acquisition:
- Place the whole explant or fixed section on a microscope slide.
- Using the GFP filter set, visualize the fluorescence. Successful infection is indicated by bright green fluorescence at the cotyledon node and in vascular tissues.
- Capture images for documentation and quantification. Compare against negative controls (e.g., explants treated with Agrobacterium lacking the GFP vector) to account for autofluorescence.
- Efficiency Calculation:
- Count the total number of treated explants and the number showing clear GFP fluorescence.
- Calculate the percentage of GFP-positive explants to determine the infection efficiency.
Advanced Image Analysis with TrueSpot Software
For high-throughput or highly quantitative studies, manual image analysis can be a bottleneck. Automated tools like TrueSpot can enhance objectivity and throughput.
Procedure:
- Image Processing: Export your fluorescence microscopy images in a standard format (e.g., TIFF).
- Software Analysis:
- Use the TrueSpot software, an automated tool designed for robust detection and quantification of fluorescent puncta in both 2D and 3D images [33].
- TrueSpot automates the challenging step of setting a signal threshold, which distinguishes true GFP signals from background noise, thereby reducing subjectivity and improving consistency across large datasets [33].
- Data Output: The software provides quantitative data on the number, intensity, and location of GFP signals, offering a more granular view of infection efficiency.
Workflow Visualization
The following diagram illustrates the logical sequence and decision-making process for using GFP fluorescence to confirm infection, from preparation to final analysis.
The Scientist's Toolkit
A successful infection assay relies on specific reagents and tools. The following table outlines essential solutions and their functions for this application.
Table 2: Essential Research Reagent Solutions for GFP-Based Infection Assay
| Reagent / Tool | Function / Purpose | Application Notes |
|---|---|---|
| pTRV2-GFP Vector | Serves as the visual reporter construct; expression confirms successful T-DNA transfer. | A component of the TRV-VIGS system; co-delivered with pTRV1 for viral replication [1]. |
| Agrobacterium tumefaciens GV3101 | The delivery vehicle for genetically introducing the GFP construct into plant cells. | A disarmed strain commonly used for plant transformation; requires specific culture conditions [1]. |
| Acetosyringone | A phenolic compound that induces the Agrobacterium Vir genes, enhancing T-DNA transfer efficiency. | Added to the Agrobacterium infection medium prior to co-cultivation [1]. |
| Paraformaldehyde (PFA) 4% | A cross-linking fixative that preserves cellular structure and prevents GFP from leaking out of cells. | Critical for unfixed tissue sections; use pre-warmed (30-37°C) for best results in post-fixation [32]. |
| TrueSpot Software | Automated, unbiased tool for detecting and quantifying fluorescent spots in microscopy images. | Outperforms other tools in precision, especially with varying background noise [33]. |
| Fluorescence Microscope | Essential equipment for exciting GFP and detecting its emitted fluorescence. | Requires a compatible GFP filter set (Ex/Em ~470/525 nm). A confocal microscope can provide better spatial resolution. |
Maximizing Silencing Efficiency: Key Parameters and Troubleshooting Common Pitfalls
Within the broader scope of a thesis investigating Virus-Induced Gene Silencing (VIGS) delivery in soybean, optimizing Agroinfiltration is a critical step for achieving high efficiency in transient gene expression and silencing. This protocol details the precise optimization of two fundamental parameters: the optical density of the Agrobacterium culture at 600 nm (OD600), which determines the bacterial concentration, and the concentration of the phenolic elicitor acetosyringone. These factors directly influence the efficiency of T-DNA transfer and the subsequent expression of the VIGS construct in soybean cotyledon nodes. The methods outlined here are adapted from established soybean transformation and VIGS protocols to create a specialized, optimized pipeline for cotyledon node VIGS delivery [1] [34] [35].
Key Parameter Optimization
Systematic optimization of physical and chemical parameters is essential for maximizing T-DNA delivery. The following tables summarize optimized values for OD600 and acetosyringone, alongside other influential factors.
Table 1: Optimized Core Parameters for Soybean Cotyledon Node Agroinfiltration
| Parameter | Optimized Concentration / Value | Experimental Context / Notes |
|---|---|---|
| Acetosyringone | 150 - 200 µM | Added to both bacterial suspension and co-cultivation media [1] [35] [36]. |
| Bacterial OD600 | 0.3 - 1.0 | Strain-dependent; lower end (0.3) for A. rhizogenes R1000 [35], higher end (1.0) for A. tumefaciens GV3101 in peony [37]. |
| Bacterial Strain | A. tumefaciens GV3101, EHA105; A. rhizogenes R1000 | GV3101 used in soybean TRV-VIGS [1]; R1000 optimal for hairy root transformation [35]. |
| Co-cultivation Time | 3 days | Standard duration under dark conditions [1] [34]. |
Table 2: Summary of Additional Optimized Factors
| Factor | Optimized Condition | Impact / Function |
|---|---|---|
| Physical Wounding | Sonication (10 min) & Vacuum Infiltration (10 min) | Significantly enhances transformation efficiency by creating micro-wounds for bacterial entry [34]. |
| Antioxidants | Lipoic acid (5 µM) [38] or Ascorbate Acid (0.56 mM) [39] | Reduces oxidative stress and tissue necrosis, improving cell viability and transformation. |
| Surfactants | Pluronic F-68 (0.002%) [38] or Tween-20 (0.03% v/v) [39] | Lowers surface tension, promoting even infiltration of the bacterial suspension into plant tissues. |
| Temperature | 37°C heat shock 1-2 days post-infiltration | Dramatically increases recombinant protein levels in transient expression [38]. |
Detailed Experimental Protocols
Protocol A: Agrobacterium Preparation and Inoculum Standardization
This procedure ensures a standardized and vir-induced bacterial culture for infiltration.
- Strain and Vector: Use Agrobacterium tumefaciens GV3101 harboring the pTRV1 and pTRV2-VIGS vectors [1].
- Starter Culture: Inoculate a single colony into 30 mL of LB broth with appropriate antibiotics (e.g., kanamycin, rifampicin). Incubate at 28°C for 16 hours with shaking at 180 rpm [34].
- Pellet and Resuspend: Centrifuge the culture at 6000 rpm for 15 minutes. Discard the supernatant.
- Induction Medium: Resuspend the bacterial pellet in a liquid inoculation medium (e.g., ½ strength MS or B5 medium) supplemented with 200 µM acetosyringone [1] [36].
- Induction Incubation: Incubate the resuspended culture for 1 hour at 28°C with gentle shaking (180 rpm) to fully activate the vir genes [34].
- OD600 Standardization: Adjust the final OD600 of the bacterial suspension to the target value (e.g., 1.0 for cotyledon node immersion [1] or 0.3 for specific hairy root assays [35]) using the same induction medium.
Protocol B: Soybean Cotyledon Node Explant Preparation and Agroinfiltration
This protocol outlines the preparation and transformation of the primary explant.
- Seed Sterilization: Surface-sterilize soybean seeds (e.g., cv. Tianlong 1 or Williams 82) with 70% ethanol for 5 minutes, followed by 4% sodium hypochlorite for 14 minutes. Rinse thoroughly with sterile distilled water [36].
- Imbibition and Dissection: Imbibe the sterilized seeds in sterile water for 24 hours. Remove the seed coat and carefully separate the cotyledons. The explant consists of the cotyledonary node with a segment of the hypocotyl attached ("modified half-seed explant") [1] [34].
- Wounding (Optional but Recommended): Subject the explants to sonication for 10 minutes in the prepared Agrobacterium suspension. This can be followed by vacuum infiltration for 10 minutes to significantly enhance transformation efficiency [34].
- Agroinfiltration: Immerse the fresh, wounded explants in the standardized Agrobacterium suspension for 20-30 minutes. Ensure complete submersion and gentle agitation [1].
- Co-cultivation: Blot-dry the explants and transfer them to a co-cultivation medium (e.g., MS medium without antibiotics, supplemented with 200 µM acetosyringone). Maintain the explants in the dark at 25 ± 2°C for 3 days [1] [34].
Workflow and Parameter Optimization Logic
The diagram below illustrates the experimental workflow and the logical decision points for parameter optimization.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Agroinfiltration
| Reagent | Function / Application | Example Usage / Concentration |
|---|---|---|
| Acetosyringone | Phenolic elicitor that activates Agrobacterium vir genes, crucial for T-DNA transfer. | 150-200 µM in bacterial suspension and co-cultivation media [1] [35]. |
| Antioxidants (Lipoic Acid, Ascorbate Acid) | Scavenge reactive oxygen species (ROS) produced during infection, reducing tissue necrosis and improving transformation efficiency. | 5 µM Lipoic Acid [38] or 0.56 mM Ascorbate Acid [39] in infiltration media. |
| Surfactants (Pluronic F-68, Tween-20) | Reduce surface tension, ensuring even and thorough infiltration of the bacterial suspension into intercellular spaces. | 0.002% Pluronic F-68 [38] or 0.03% Tween-20 [39]. |
| Agrobacterium Strains | Vehicle for T-DNA delivery. Strain choice impacts host range and transformation efficiency. | GV3101 for TRV-VIGS [1]; R1000 for hairy root transformation [35]. |
| TRV-based VIGS Vectors | Viral vectors (pTRV1, pTRV2) engineered to carry host gene fragments for targeted gene silencing. | pTRV2-GFP derivatives used to silence endogenous genes like GmPDS [1]. |
| 5-Azacytidine (AzaC) | DNA methyltransferase inhibitor; can increase transgene expression by reducing DNA methylation of the transferred T-DNA. | 20 µM in infiltration media [39]. |
Within the context of cotyledon node Virus-Induced Gene Silencing (VIGS) research in soybean, precise control of environmental conditions is not merely a matter of optimal plant growth—it is a fundamental determinant of experimental success and reproducibility. Soybean (Glycine max L.), a quantitative short-day plant, exhibits profound sensitivity to photoperiod and temperature, which directly influences its developmental trajectory and the efficiency of viral vectors used in functional genomics studies [40]. The establishment of a robust VIGS system, particularly one utilizing Agrobacterium-mediated infection of cotyledon nodes, requires careful optimization of these factors to ensure high infection rates, potent systemic silencing, and accurate phenotypic interpretation [1] [7]. This application note details the critical environmental parameters and provides standardized protocols to support researchers in generating consistent and reliable data in soybean VIGS experiments.
Quantitative Data on Environmental Factors
The following tables consolidate key quantitative data on the influence of photoperiod and temperature on soybean development and VIGS efficiency, providing a reference for experimental design.
Table 1: Impact of Photoperiod on Soybean Developmental Rate to Flowering (R1) [40]
| Photoperiod (Hours) | Impact on Time to Flowering (R1) | Impact on Node Appearance Rate (NdAR) |
|---|---|---|
| 14 h | Faster flowering | Highest NdAR (0.021 nodes per hour of photoperiod) |
| 15 h | Delayed flowering | Optimal NdAR during early development (VE to VC) |
| 16 h | Delayed flowering | Lower NdAR |
| 17 h | Most significantly delayed flowering | Lowest NdAR (0.016 nodes per hour of photoperiod) |
Table 2: Optimized Environmental Conditions for Cotyledon Node VIGS in Soybean [1] [7] [25]
| Environmental Factor | Recommended Setting | Experimental Impact and Rationale |
|---|---|---|
| Pre-inoculation Growth | 16 h light / 8 h dark, 23°C | Standard conditions for consistent seedling development prior to Agrobacterium infection [25]. |
| Post-inoculation Growth | 16 h light / 8 h dark, 23°C | Maintained to support plant health and silencing development; TRV-based VIGS is effective under these conditions [1] [25]. |
| Agroinfiltration Temperature | 22-25°C | Typical room temperature range for the Agrobacterium incubation step [1]. |
| Photoperiod for Speed Breeding | 22 h post-flowering initiation | Applied to accelerate generation cycling; note this is a specialized protocol for breeding, not standard VIGS [41]. |
Experimental Protocols for Environmental Control in VIGS
Protocol: Establishing Baseline Plant Growth for Cotyledon Node VIGS
This protocol ensures uniform plant material prior to Agrobacterium infection.
- Seed Sterilization and Germination: Surface-sterilize soybean seeds (e.g., cultivar Tianlong 1 or Aram) and soak in sterile water until swollen. Bisect seeds longitudinally to obtain half-seed explants [1].
- Pre-conditioning Growth: Place explants or germinated seedlings in a growth chamber programmed to 16 hours of light and 8 hours of dark at a constant temperature of 23-25°C [25] [1].
- Light Quality: Utilize cool white LED lights with a photosynthetic photon flux of 550 μmol/(m²s) at canopy level. Supplementation with red light (660 nm) can be beneficial for later developmental stages [41].
- Duration: Grow plants under these conditions until the cotyledons are fully expanded and the first emerging leaves are visible, typically for 2 weeks post-germination [25].
Protocol: Agrobacterium-Mediated VIGS via Cotyledon Node and Post-Inoculation Care
This optimized protocol leverages the cotyledon node for high-efficiency delivery of TRV-based VIGS constructs [1].
- Vector Construction: Clone a 200-400 bp fragment of the target gene (e.g., GmPDS) into the multiple cloning site of a pTRV2 vector [1] [42].
- Agrobacterium Preparation:
- Transform recombinant pTRV2 and the helper pTRV1 plasmids separately into Agrobacterium tumefaciens strain GV3101.
- Grow individual bacterial cultures in LB medium with appropriate antibiotics at 28°C for 12-16 hours.
- Harvest cells by centrifugation and resuspend in an infiltration medium (10 mM MES pH 5.6, 10 mM MgCl₂, and 150 μM acetosyringone) to a final optical density at 600 nm (OD₆₀₀) of 0.8-1.0 [1] [25].
- Mix the pTRV1 and pTRV2 cultures in a 1:1 ratio and incubate at room temperature for 3-4 hours before use.
- Cotyledon Node Infection:
- Immerse the prepared fresh half-seed explants in the Agrobacterium suspension for 20-30 minutes, ensuring the cotyledonary node is fully exposed to the solution [1].
- Alternative Agroinfiltration Method: For some genotypes, direct injection or vacuum infiltration of the Agrobacterium suspension into cotyledon nodes may be used, though immersion is reported to be more efficient for soybean [1] [8].
- Post-Inoculation Incubation and Plant Growth:
- Co-cultivate the infected explants on sterile filter paper in the dark at 22-25°C for 2-3 days.
- Transfer plants to soil or tissue culture medium and return them to the growth chamber set at 16 hours light / 8 hours dark and 23°C [25].
- Maintain high humidity for the first few days after transfer to reduce transplant stress.
- Phenotype Monitoring: Observe plants for systemic silencing phenotypes, such as photobleaching for GmPDS, which typically becomes visible in newly developed leaves and cluster buds around 21 days post-inoculation (dpi) [1].
Signaling Pathways and Experimental Workflows
The following diagrams illustrate the molecular mechanism of VIGS and the integrated experimental workflow that incorporates critical environmental parameters.
Diagram 1: VIGS mechanism and environmental influence.
Diagram 2: Cotyledon node VIGS workflow.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Soybean Cotyledon Node VIGS
| Item | Function / Application | Specific Examples / Notes |
|---|---|---|
| VIGS Vector System | Delivers target gene sequence to trigger host RNAi. | Tobacco Rattle Virus (TRV): pTRV1 (helper plasmid) and pTRV2 (with MCS for insert) are most versatile [1] [7]. |
| Agrobacterium tumefaciens Strain | Mediates vector delivery into plant cells. | GV3101: Commonly used, disarmed strain with high transformation efficiency for soybean cotyledon nodes [1] [25]. |
| Soybean Genotype | Host plant for functional gene validation. | Tianlong 1, Aram: Cultivars demonstrated to have high VIGS efficiency (65-95%) and/or tolerance to viral symptoms [1] [25]. |
| Infiltration Medium | Suspension medium for Agrobacterium during infection. | 10 mM MES (pH 5.6), 10 mM MgCl₂, 150 μM Acetosyringone; induces Vir genes for T-DNA transfer [1] [25]. |
| Selection Antibiotics | Maintains plasmid integrity in bacterial and plant cultures. | Kanamycin, Rifampicin; specific antibiotics depend on the resistance markers of the vectors and Agrobacterium strain. |
| Positive Control Silencing Construct | Validates the VIGS system is functioning. | pTRV2-GmPDS: Silencing Phytoene Desaturase causes photobleaching, a visual marker for success [1] [25]. |
| Growth Chamber | Provides controlled photoperiod and temperature. | Programmable for 16h light / 8h dark at 23-25°C; consistent conditions are vital for reproducibility [1] [25]. |
In soybean (Glycine max L.) research, virus-induced gene silencing (VIGS) has emerged as a powerful reverse genetics tool for rapid functional validation of candidate genes. However, its application has been significantly hampered by the formidable physical barriers presented by the plant's aerial surfaces—specifically, thick cuticles and dense trichomes. These structures inherently protect the plant from environmental stresses and pathogen ingress, but consequently impede efficient Agrobacterium-mediated delivery of VIGS vectors.
Conventional infection methods, such as leaf misting or direct injection, often prove inadequate against these barriers. The dense trichome layer and waxy cuticle of soybean leaves physically block liquid penetration, while the thick cuticular layer more broadly restricts the entry of Agrobacterium, a vehicle for Tobacco Rattle Virus (TRV) vectors [1]. This results in unacceptably low transformation efficiencies, creating a critical bottleneck for high-throughput functional genomics. This Application Note details an optimized, tissue culture-based protocol that systematically overcomes these barriers by targeting the more accessible cotyledon node with an immersion-based delivery system.
Comparative Analysis of Infection Methods
The table below summarizes the key limitations of conventional methods compared to the optimized cotyledon node immersion protocol for achieving efficient VIGS in soybean.
Table 1: Comparison of VIGS Delivery Methods in Soybean
| Method | Key Procedure | Reported Efficiency | Major Limitations |
|---|---|---|---|
| Conventional (Misting/Injection) | Spraying or infiltrating Agrobacterium suspension onto leaves | Low (Not quantified) | Thick cuticle and dense trichomes impede liquid penetration [1] |
| Optimized Cotyledon Node Immersion | Using bisected half-seed explants; immersion in Agrobacterium suspension for 20-30 min | 65% - 95% (by qPCR) | Requires sterile tissue culture conditions and explant preparation [1] |
Optimized Protocol for Cotyledon Node VIGS Delivery
This section provides a detailed, step-by-step methodology for the efficient delivery of TRV-based VIGS constructs into soybean via the cotyledon node.
Key Research Reagent Solutions
The following table lists the essential reagents and materials required for the successful implementation of this protocol.
Table 2: Essential Reagents and Materials for Soybean Cotyledon Node VIGS
| Item | Specification / Example | Function / Purpose |
|---|---|---|
| VIGS Vector | pTRV1, pTRV2-GFP derivatives (e.g., pTRV:GmPDS) [1] | TRV-based silencing vector system |
| Agrobacterium Strain | GV3101 [1] | Delivery of recombinant VIGS vectors |
| Plant Material | Sterilized soybean seeds (e.g., cv. Tianlong 1) [1] | Source of half-seed explants |
| Antibiotics | Kanamycin, Rifampicin [1] | Selection for vector and Agrobacterium strain |
| Induction Agent | Acetosyringone | Induction of Agrobacterium virulence genes |
| Surface Sterilant | Sodium hypochlorite, Ethanol | Seed surface sterilization |
Step-by-Step Experimental Procedure
Vector Construction and Agrobacterium Preparation
- Clone the target gene fragment (e.g., GmPDS, ~300-500 bp) into the pTRV2 vector using appropriate restriction enzymes (e.g., EcoRI and XhoI) [1].
- Introduce the recombinant pTRV2 and helper pTRV1 plasmids into Agrobacterium tumefaciens GV3101 via electroporation or freeze-thaw method.
- Culture positive Agrobacterium clones in LB medium supplemented with appropriate antibiotics (e.g., Kanamycin, Rifampicin) at 28°C for 24-48 hours.
Explant Preparation
- Surface-sterilize soybean seeds using a sterilant solution.
- Soak the sterilized seeds in sterile water until they are fully swollen.
- Aseptically bisect the swollen seeds longitudinally to obtain half-seed explants, ensuring the cotyledon node is intact [1].
Agro-infection via Immersion
- Harvest the Agrobacterium cells by centrifugation and resuspend them in an induction medium (e.g., MES buffer with acetosyringone) to an optimal optical density (OD₆₀₀ of ~1.0).
- Immerse the fresh half-seed explants in the Agrobacterium suspension for 20-30 minutes with gentle agitation. This duration has been identified as optimal for efficient infection [1].
- Briefly blot the explants on sterile paper to remove excess liquid.
Co-cultivation and Plant Regeneration
- Transfer the infected explants to co-cultivation medium and incubate in the dark at 22-25°C for 2-3 days.
- After co-cultivation, transfer the explants to a regeneration medium containing antibiotics to suppress Agrobacterium overgrowth and select for transformed tissues.
- Maintain the plants under standard growth chamber conditions (e.g., 16/8 h light/dark cycle).
Workflow Visualization and Validation
The following diagram illustrates the complete experimental workflow, from explant preparation to the validation of successful gene silencing.
Diagram 1: Soybean Cotyledon Node VIGS Workflow
Validation of Infection and Silencing Efficiency
Infection Efficiency: Around the fourth day post-infection, a portion of the hypocotyl can be excised and observed under a fluorescence microscope for GFP signals. Successful infection is indicated by fluorescence in 2-3 cell layers initially, spreading to deeper tissues, with effective infectivity efficiency exceeding 80% and reaching up to 95% for certain cultivars like Tianlong 1 [1]. Transverse sections should show over 80% of cells with successful infiltration [1].
Silencing Phenotype and Efficiency: For a positive control like GmPDS, photobleaching in leaves inoculated with pTRV:GmPDS typically becomes visible at approximately 21 days post-inoculation (dpi), starting in the cluster buds, while no such phenotype is observed in empty vector controls [1]. Silencing efficiency should be quantified using qPCR, which has confirmed a range of 65% to 95% for this optimized protocol [1].
Concluding Remarks
The thick cuticle and dense trichomes of soybean represent a significant but surmountable obstacle for VIGS delivery. The optimized protocol detailed herein, which employs a cotyledon node explant and a critical immersion step, directly counteracts these physical barriers. This method achieves a dramatic increase in infection efficiency, transforming VIGS into a reliable and powerful tool for the rapid functional characterization of genes in soybean. By enabling high-efficiency, systemic silencing, this approach will significantly accelerate research in disease resistance, stress tolerance, and overall soybean functional genomics.
Within the context of advancing cotyledon node-based VIGS delivery in soybean research, the precise monitoring of the timeline from viral infection to the manifestation of a visible silencing phenotype is paramount. Virus-Induced Gene Silencing (VIGS) serves as a powerful functional genomics tool to rapidly characterize gene function, particularly in crops like soybean where stable transformation is time-consuming and challenging [1] [14]. The establishment of a tobacco rattle virus (TRV)-based system, optimized for delivery via the cotyledon node, has significantly improved the efficiency of this process in soybean [1] [43]. This application note delineates a detailed protocol and monitoring schedule for effective VIGS experimentation, with systemic silencing and clear phenotypic outcomes typically observable by 21 days post-inoculation (dpi).
Materials and Reagent Solutions
The following reagents are essential for the successful implementation of the cotyledon node VIGS protocol in soybean.
Table 1: Key Research Reagent Solutions for Cotyledon Node VIGS in Soybean
| Reagent/Vector Name | Type/Function | Key Application in Protocol |
|---|---|---|
| pTRV1 and pTRV2 Vectors | Tobacco Rattle Virus (TRV)-based VIGS vectors [1] | pTRV1 encodes viral replication proteins; pTRV2 carries the target gene fragment for silencing. |
| Agrobacterium tumefaciens GV3101 | Disarmed Agrobacterium strain [1] | Acts as the delivery vehicle for the TRV vectors into plant cells via cotyledon node infiltration. |
| Phytoene Desaturase (GmPDS) | Endogenous marker gene [1] | Silencing causes photobleaching, providing a visual, non-lethal marker for assessing VIGS efficiency. |
| Soybean Cultivar 'Tianlong 1' | Plant Material [1] | A soybean genotype demonstrated to be effective for this protocol, with high infection efficiency. |
Experimental Protocol
Vector Construction and Agrobacterium Preparation
- Clone Target Gene Fragment: Amplify a 200-400 bp fragment from the coding sequence (CDS) of the target soybean gene (e.g., GmPDS, GmRpp6907) [1] [12].
- Insert into TRV2 Vector: Ligate the purified PCR product into the multiple cloning site of the pTRV2-GFP vector, downstream of the coat protein promoter, using appropriate restriction enzymes (e.g., EcoRI and XhoI) [1].
- Transform Agrobacterium: Introduce the recombinant pTRV2 construct and the separate pTRV1 construct into Agrobacterium tumefaciens strain GV3101 via electroporation or freeze-thaw transformation [1].
- Prepare Agrobacterium Culture: Inoculate single colonies of Agrobacterium containing pTRV1 and the recombinant pTRV2 into separate liquid culture media with appropriate antibiotics. Grow cultures at 28°C with shaking until they reach an OD₆₀₀ of 0.4-0.6 [1].
- Induce Bacterial Cells: Pellet the bacterial cultures by centrifugation and resuspend them in an induction medium (e.g., containing 10 mM MES, 10 mM MgCl₂, and 200 µM acetosyringone) to activate the Vir genes. Incubate the suspensions for 3-4 hours at room temperature [1].
- Prepare Inoculum Mix: Combine the induced pTRV1 and pTRV2 cultures in a 1:1 ratio. Adjust the final OD₆₀₀ of the mixture to approximately 0.8 for inoculation [1].
Cotyledon Node Agroinfiltration
- Plant Material Preparation: Surface-sterilize soybean seeds and soak them in sterile water until they swell. Carefully bisect the soaked seeds longitudinally to create half-seed explants, ensuring the cotyledon node is intact and exposed [1].
- Agroinfiltration: Immerse the fresh half-seed explants in the prepared Agrobacterium inoculum mix. Ensure the cotyledon node is fully submerged. Perform the immersion for 20-30 minutes with gentle agitation to facilitate optimal bacterial infection [1].
- Co-cultivation and Plant Growth: After infiltration, blot the explants dry on sterile filter paper and transfer them to a sterile tissue culture system for co-cultivation. Maintain the plants under standard growth conditions (e.g., 25°C day/20°C night, 16-hour photoperiod) [1].
Timeline and Phenotype Monitoring
Systemic monitoring of gene silencing is critical for phenotypic assessment. The table below outlines the key milestones from infection to visible silencing.
Table 2: Quantitative Monitoring Timeline for VIGS in Soybean (Key Milestones)
| Days Post-Inoculation (dpi) | Monitoring Event | Expected Observation / Quantitative Measure |
|---|---|---|
| 4 dpi | Initial Infection Efficiency | Fluorescence observed in >80% of cotyledon node cells under microscope [1]. |
| 14 dpi | Early Systemic Silencing | Onset of target gene mRNA downregulation detectable via qPCR in newly emerged leaves [1]. |
| 21 dpi | Visible Phenotype Manifestation | Clear photobleaching in leaves (for GmPDS silencing); silencing efficiency reaches 65-95% [1]. |
| 28 dpi & Beyond | Phenotype Stability & Analysis | Stable silencing phenotype allows for robust functional characterization of target genes (e.g., disease susceptibility) [1]. ``` |
Diagram 1: VIGS Experimental Workflow from Preparation to Phenotype Analysis.
Data Analysis and Interpretation
Quantitative Assessment of Silencing Efficiency
The success of the VIGS protocol is quantitatively evaluated through molecular and phenotypic analyses.
Table 3: Key Metrics for Evaluating VIGS Efficiency at 21 dpi
| Analysis Method | Parameter Measured | Expected Outcome in Successfully Silenced Plants |
|---|---|---|
| qRT-PCR | Relative mRNA abundance of target gene | 65-95% reduction in transcript levels compared to empty vector controls [1]. |
| Visual Phenotyping | Presence of tissue bleaching (GmPDS) or disease susceptibility | Clear, systemic photobleaching in leaves and meristems [1]. |
| Fluorescence Microscopy | GFP signal in cotyledon nodes (at 4 dpi) | Infection efficiency indicator; >80% of cells show fluorescence [1]. |
Troubleshooting Common Issues
- Low Infection Efficiency: Ensure the cotyledon node is not damaged during explant preparation and is fully submerged during agroinfiltration. Verify the OD₆₀₀ and induction of the Agrobacterium culture [1].
- No Visible Phenotype at 21 dpi: Confirm the integrity and specificity of the insert in the pTRV2 vector by sequencing. Check plant growth conditions, as temperature and light intensity can influence the rate of viral spread and silencing [1].
- Unsynchronized Silencing: The use of half-seed explants from uniformly sterilized and pre-swollen seeds promotes synchronized infection, leading to more consistent silencing across a plant population [1].
Diagram 2: Molecular Mechanism of Virus-Induced Gene Silencing.
Application Notes
- Critical Step: The cotyledon node infiltration method is crucial for overcoming the physical barriers posed by the thick cuticle and dense trichomes of soybean leaves, which impede other inoculation methods [1].
- Positive Control: Always include a pTRV2-GmPDS construct as a positive control. The appearance of photobleaching by 21 dpi confirms the entire system is functioning optimally [1].
- Experimental Design: Employ appropriate negative controls, such as plants inoculated with the pTRV:empty vector (pTRV1 + pTRV2-GFP without a target insert), to distinguish virus-induced symptoms from true gene silencing phenotypes [1].
- Utility in Functional Genomics: This protocol has been successfully applied to validate the function of key genes in soybean disease resistance, such as the rust resistance gene GmRpp6907 and the defense-related gene GmRPT4, demonstrating its robustness for rapid gene characterization [1].
Virus-Induced Gene Silencing (VIGS) is a powerful tool for rapid functional genomics in soybean, a vital crop for global food security [1]. The cotyledon node delivery method has emerged as an efficient technique for implementing VIGS in soybean research [1] [31]. However, the reliability of findings generated through this approach critically depends on ensuring the specificity of silencing effects. Proper experimental controls and strategies to minimize off-target effects are fundamental to distinguishing specific gene silencing phenotypes from artifacts caused by the viral vector or non-targeted gene silencing. This application note provides detailed protocols and frameworks for maintaining specificity in cotyledon node-based VIGS experiments in soybean, enabling researchers to generate robust, interpretable data for gene function characterization.
Core Principles of VIGS Specificity
The biological foundation of VIGS lies in the plant's post-transcriptional gene silencing (PTGS) machinery, an antiviral defense mechanism [7]. When a recombinant viral vector carrying a fragment of a plant gene infects the plant, double-stranded RNA replication intermediates are processed by Dicer-like (DCL) enzymes into 21-24 nucleotide small interfering RNAs (siRNAs). These siRNAs are incorporated into the RNA-induced silencing complex (RISC), which guides sequence-specific degradation of complementary endogenous mRNA transcripts [7]. This same mechanism can inadvertently lead to off-target effects if the siRNA populations silence genes with sufficient sequence similarity to the target fragment.
Essential Experimental Controls for Cotyledon Node VIGS
Implementing a complete set of experimental controls is the primary strategy for verifying that observed phenotypes result specifically from silencing the target gene. The table below summarizes the essential controls for a rigorous VIGS experiment.
Table 1: Essential Experimental Controls for Soybean Cotyledon Node VIGS
| Control Type | Description | Purpose | Interpretation of Results |
|---|---|---|---|
| Empty Vector Control | Plants inoculated with TRV vectors (pTRV1 + pTRV2) lacking any plant gene insert [1]. | Accounts for physiological effects caused by viral infection and Agrobacterium infiltration. | Any phenotype observed in test plants but not in this control is likely due to specific gene silencing. |
| Visual Marker Control | Plants inoculated with a vector targeting a gene with a known visible phenotype (e.g., GmPDS causing photobleaching) [1] [31]. | Validates the entire VIGS protocol is working efficiently in the experimental conditions. | Successful photobleaching indicates efficient transfection, viral spread, and silencing. |
| Multiple Independent Target Fragments | Designing two or more non-overlapping VIGS constructs targeting different regions of the same gene [7]. | Confirms phenotype specificity. | Similar phenotypes from multiple independent fragments provide strong evidence for correct gene function assignment. |
| Wild-type / Untreated Control | Untreated plants or plants infiltrated with an Agrobacterium suspension lacking the TRV vector. | Monitors natural development and rules out effects from growth conditions. | Serves as a baseline for comparing plant growth and morphology. |
Strategies to Minimize Off-Target Effects
Off-target effects occur when siRNAs derived from the VIGS construct silence non-target genes due to sequence similarity. The following strategies are critical for mitigating this risk.
Careful Insert Design and In Silico Analysis
- Fragment Length and Position: Use inserts of 200-400 base pairs from the coding sequence of the target gene [31] [7].
- Sequence Specificity Verification: Before cloning, perform a BLASTN search of the chosen fragment against the soybean genome to identify and avoid sequences with high similarity (e.g., >80-85% identity over >50 bp) to other genes [31] [7].
- Avoiding Conserved Domains: When possible, target unique regions of the gene, such as the 3' or 5' untranslated regions (UTRs), rather than highly conserved functional domains shared among gene family members.
Silencing Efficiency and Phenotype Correlation
The correlation between the level of target gene transcript reduction and the severity of the phenotype is a strong indicator of specificity. Quantitative data from studies using cotyledon node VIGS demonstrate this relationship.
Table 2: Quantitative Silencing Efficiency and Phenotypic Correlation in Soybean VIGS
| Silenced Gene | Gene Function | Silencing Efficiency (qPCR) | Observed Phenotype | Citation |
|---|---|---|---|---|
| GmPDS | Carotenoid biosynthesis | Not specified | Systemic photobleaching in leaves | [1] |
| GmRpp6907 | Rust resistance | Not specified | Compromised rust immunity | [1] |
| GmRPT4 | Defense response | Not specified | Altered defense responses | [1] |
| NcChlH | Chlorophyll synthesis | Up to 84.4% | Chlorosis in true leaves | [31] |
Detailed Protocol: A Specificity-Focused Workflow
The following diagram and protocol outline the key steps for a cotyledon node VIGS experiment in soybean, with integrated steps for ensuring specificity.
Stage 1: Vector Construction and Specificity Checks
- Insert Selection: Design primers to amplify a 250-350 bp fragment from the target gene's cDNA. Follow the principles outlined in Section 4.1.
- Bioinformatic Control: Use the BLAST tool against the Glycine max genome database to screen the selected fragment for potential off-targets.
- Cloning: Ligate the purified PCR product into the pTRV2 vector digested with appropriate restriction enzymes (e.g., EcoRI and XhoI) [1]. Transform the construct into E. coli DH5α and verify the sequence of positive clones.
- Agrobacterium Preparation: Introduce the verified plasmid and the pTRV1 plasmid into Agrobacterium tumefaciens strain GV3101 separately using the freeze-thaw method [31].
Stage 2: Plant Inoculation and Control Monitoring
- Agro-inoculum Preparation:
- Grow individual Agrobacterium cultures containing pTRV1 and your pTRV2-derivatives (e.g., pTRV2:empty, pTRV2:GmPDS, pTRV2:YourGene) in LB medium with appropriate antibiotics at 28°C for 24-48 hours.
- Pellet the bacteria by centrifugation and resuspend in an infiltration buffer (10 mM MgCl₂, 10 mM MES, 100 µM acetosyringone) to a final OD600 of 1.0-2.0 [1] [23].
- Mix the pTRV1 and each pTRV2 suspension in a 1:1 ratio and incubate at room temperature for 3-4 hours before infiltration.
- Cotyledon Node Infiltration:
- Surface-sterilize soybean seeds and allow them to imbibe water until swollen.
- Prepare half-seed explants by longitudinally bisecting the swollen seeds.
- Immerse the fresh cotyledon node explants in the Agrobacterium suspension for 20-30 minutes, ensuring full contact [1].
- Alternatively, use a syringe without a needle to apply the suspension directly to the cotyledon node.
- Plant Growth and Control Monitoring:
- Transplant the treated explants to soil or sterile medium and grow under standard conditions (e.g., 16/8 hour light/dark photoperiod, 25°C).
- Closely monitor plants inoculated with the visual marker control (e.g., TRV2:GmPDS). The appearance of photobleaching 2-4 weeks post-inoculation confirms a successful experiment [1] [31].
- Simultaneously, document the health and morphology of the empty vector control plants.
Stage 3: Molecular Validation of Specificity
- RNA Extraction: From silenced tissue (showing phenotype) and control tissue, extract total RNA using a commercial plant RNA kit.
- Quantitative RT-PCR (qRT-PCR) for Target Gene:
- Synthesize cDNA from the extracted RNA.
- Perform qRT-PCR with primers designed to amplify a region of the target gene outside the sequence used for the VIGS construct.
- Use soybean housekeeping genes (e.g., Ubiquitin, Actin) for normalization. A significant reduction (e.g., >50%) in transcript levels in test plants compared to empty vector controls confirms silencing [31].
- qRT-PCR for Off-Target Genes:
- Based on the initial BLAST analysis, select 3-5 genes with the highest sequence similarity to your VIGS insert.
- Design qRT-PCR primers for these candidate off-target genes.
- Perform qRT-PCR as above. The absence of significant transcript reduction in these genes strengthens the claim of a specific silencing effect.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Research Reagents for Soybean Cotyledon Node VIGS
| Reagent / Material | Function / Purpose | Example / Specification |
|---|---|---|
| TRV Vectors | Bipartite viral vector system for VIGS; TRV1 encodes replication proteins, TRV2 carries the target gene insert. | pTRV1, pTRV2 [1] [7] |
| Agrobacterium tumefaciens | Bacterial strain used to deliver the TRV DNA vectors into plant cells. | Strain GV3101 [1] [31] |
| Infiltration Buffer | Solution for suspending and activating Agrobacterium for plant infiltration. | 10 mM MgCl₂, 10 mM MES (pH 5.7), 100 µM acetosyringone [23] |
| Visual Marker Construct | Positive control vector to visually confirm VIGS efficiency. | pTRV2-GmPDS (soybean) or pTRV2-ChlH (catmint) [1] [31] |
| Restriction Enzymes | For directional cloning of the target fragment into the TRV2 vector. | EcoRI, XhoI, BamHI [1] [31] |
| qRT-PCR Reagents | For molecular validation of target gene silencing and specificity checks. | SYBR Green Master Mix, gene-specific primers [1] [31] |
Validation, Applications, and Comparative Analysis of the Cotyledon Node VIGS System
Virus-induced gene silencing (VIGS) is a powerful reverse genetics tool for rapid functional analysis of plant genes. The validation of any VIGS system requires a visual marker to demonstrate successful silencing, for which phytoene desaturase (PDS) has become the gold standard. Silencing PDS disrupts chlorophyll biosynthesis, resulting in a characteristic photobleaching phenotype that serves as a reliable visual indicator of effective gene silencing. This case study details the application of a cotyledon node-based VIGS delivery system to silence GmPDS in soybean (Glycine max L.), establishing an efficient platform for functional genomics in this agronomically important crop [1] [44].
Experimental Protocol
Vector Construction and Agrobacterium Preparation
The tobacco rattle virus (TRV)-based VIGS system was employed, consisting of two components: pTRV1 (encoding replication and movement proteins) and pTRV2 (carrying the target gene fragment) [1] [45].
Key Steps:
- Fragment Amplification: A 300-500 bp fragment of the GmPDS gene (GenBank accession no. M64704.1) was amplified from soybean cDNA using gene-specific primers with incorporated restriction sites (EcoRI and XhoI) [1].
- Vector Construction: The PCR-amplified GmPDS fragment was cloned into the pTRV2-GFP vector through restriction digestion and ligation. The recombinant plasmid was sequenced to confirm correct orientation and sequence integrity [1].
- Agrobacterium Transformation: The confirmed pTRV2-GmPDS plasmid and the pTRV1 plasmid were independently transformed into Agrobacterium tumefaciens strain GV3101 using heat shock or electroporation methods [1] [46].
- Culture Preparation: Single colonies of transformed Agrobacterium were inoculated in Luria-Bertani (LB) medium containing appropriate antibiotics (50 μg/ml kanamycin, 25 μg/ml rifampicin) and grown overnight at 28°C with shaking. Cultures were centrifuged and resuspended in infiltration buffer (10 mM MgCl₂, 10 mM MES, 200 μM acetosyringone, pH 5.6) to an optimal OD₆₀₀ of 1.0-2.0. The suspensions were incubated at room temperature for 2-4 hours before use [44] [46].
Plant Material and Cotyledon Node Inoculation
Soybean seeds of cultivar Tianlong 1 were surface-sterilized and germinated in sterile conditions. The optimized cotyledon node infection method was employed as follows [1]:
- Seed Preparation: Sterilized soybeans were soaked in sterile water until swollen, then longitudinally bisected to obtain half-seed explants.
- Agroinfiltration: Fresh explants were immersed in the Agrobacterium suspension containing a 1:1 mixture of pTRV1 and pTRV2-GmPDS for 20-30 minutes with gentle agitation.
- Co-cultivation: Infected explants were transferred to sterile tissue culture media and maintained under controlled conditions (23-27°C, 16/8 h light/dark photoperiod) for 3-4 days.
- Plant Development: Successfully infected plantlets were transferred to nutrient soil for further growth and development.
Validation Methods
Visual Assessment: Plants were monitored daily for the development of photobleaching symptoms, which typically appeared 14-21 days post-inoculation (dpi) [1].
Molecular Validation:
- GFP Fluorescence: Infection efficiency was assessed at 4 dpi by examining GFP fluorescence in hypocotyl sections under a fluorescence microscope [1].
- Gene Expression Analysis: RNA was extracted from silenced and control tissues. Quantitative RT-PCR was performed to measure GmPDS transcript levels using gene-specific primers [1] [46].
- Chlorophyll Content Measurement: Chlorophyll was extracted from silenced and control leaves using organic solvents, and absorbance measured spectrophotometrically to quantify chlorophyll a and b content [11].
Results and Data Analysis
Efficiency of Cotyledon Node VIGS System
Table 1: Efficiency Metrics of the Cotyledon Node VIGS System
| Parameter | Result | Measurement Method |
|---|---|---|
| Agroinfection Efficiency | >80% (up to 95% for Tianlong 1) | GFP fluorescence observation [1] |
| Silencing Onset | 14-21 days post-inoculation | Visual observation of photobleaching [1] |
| Systemic Silencing Spread | Throughout plant (leaves, stems, buds) | Phenotypic observation [1] |
| Silencing Efficiency | 65-95% | qRT-PCR analysis of GmPDS expression [1] |
GmPDS Silencing and Photobleaching Phenotype
Table 2: Characterization of GmPDS Silencing Effects
| Characteristic | Control Plants (pTRV:empty) | GmPDS-Silenced Plants |
|---|---|---|
| Leaf Color | Green | White or yellow photobleached [1] |
| Plant Vigor | Normal | Reduced growth rate [1] |
| GmPDS Expression | 100% | 5-35% of control levels [1] |
| Chlorophyll Content | Normal | Significantly reduced [11] |
| Phenotype Stability | N/A | Maintained throughout growth period [1] |
The photobleaching phenotype initially appeared in the cluster buds and systematically spread to newly developed leaves. Seedlings from the pTRV:GmPDS treatment group exhibited clear photobleaching compared to pTRV:empty controls, confirming successful systemic silencing of GmPDS [1].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Cotyledon Node VIGS
| Reagent/Solution | Function/Application | Specifications/Alternatives |
|---|---|---|
| TRV Vectors (pTRV1/pTRV2) | Viral backbone for VIGS construct | pTRV1: replication/movement proteins; pTRV2: target gene insertion [45] |
| Agrobacterium tumefaciens | Vector delivery system | Strain GV3101 with appropriate antibiotic resistance [1] [46] |
| Infiltration Buffer | Agrobacterium resuspension medium | 10 mM MgCl₂, 10 mM MES (pH 5.6), 200 μM acetosyringone [44] [46] |
| Acetosyringone | Vir gene inducer | Enhances T-DNA transfer; 100-200 μM in infiltration buffer [46] |
| Antibiotics | Selection of transformed strains | Kanamycin (50 μg/ml), Rifampicin (25-50 μg/ml) [46] |
| Plant Tissue Culture Media | Support plant growth post-infection | MS media with appropriate sucrose and vitamins [1] |
Signaling Pathways and Experimental Workflows
Discussion and Technical Notes
The cotyledon node VIGS delivery method demonstrated high efficiency (65-95% silencing) for soybean, overcoming limitations of conventional approaches like misting and direct injection, which show low infection efficiency due to soybean's thick cuticle and dense trichomes [1]. The system's effectiveness is evidenced by both the visual photobleaching phenotype and molecular confirmation of GmPDS downregulation.
Critical Success Factors:
- Plant Developmental Stage: Five-day-old seedlings with fully expanded cotyledons showed optimal silencing efficiency [11].
- Agrobacterium Density: OD₆₀₀ of 1.0-2.0 provided optimal infection without excessive stress [44].
- Infiltration Duration: 20-30 minutes immersion time balanced infection efficiency with plant viability [1].
- Environmental Conditions: Post-inoculation maintenance at 23-27°C with 16/8 h light/dark photoperiod enhanced silencing spread [44].
Troubleshooting:
- Low Silencing Efficiency: Optimize Agrobacterium density and infiltration duration; confirm fragment insertion orientation.
- Limited Systemic Spread: Ensure proper plant growth conditions; verify vector construction integrity.
- Plant Stress Symptoms: Reduce Agrobacterium density; shorten infiltration time.
This validated GmPDS silencing protocol provides a foundation for functional studies of diverse soybean genes, particularly those involved in disease resistance and stress tolerance, accelerating soybean improvement programs.
Asian Soybean Rust (ASR), caused by the obligate biotrophic fungus Phakopsora pachyrhizi, represents one of the most devastating threats to global soybean production, capable of causing yield losses ranging from 40% to as high as 90% under favorable disease conditions [47] [48]. The economic impact is particularly severe in tropical and subtropical regions, with countries like Brazil spending over $2 billion annually on ASR prevention and management, primarily through fungicide applications that pose environmental risks and face diminishing effectiveness due to emerging resistance [47]. In this context, genetic resistance offers the most sustainable and environmentally friendly approach to ASR management.
The identification of the GmRpp6907 resistance gene (originally designated Rpp6907-7) from the Chinese soybean landrace SX6907 represents a transformative breakthrough in ASR resistance breeding [47] [48]. This gene confers exceptional broad-spectrum resistance to diverse ASR populations from the United States and Brazil, making it a prime candidate for soybean improvement programs [48]. However, functional validation of resistance genes like GmRpp6907 requires robust molecular tools that can overcome the limitations of conventional genetic transformation in soybean, which remains notoriously recalcitrant, time-consuming, and genotype-dependent [1] [8].
Virus-Induced Gene Silencing (VIGS) has emerged as a powerful reverse genetics tool for rapid functional characterization of candidate genes in plants. This article details the application of an optimized Tobacco Rattle Virus (TRV)-based VIGS system for silencing GmRpp6907 in soybean, providing researchers with a high-throughput method to validate the function of this critical resistance gene within the context of cotyledon node-based delivery systems.
Technical Background
The GmRpp6907 Resistance Gene
GmRpp6907 encodes an atypical nucleotide-binding leucine-rich repeat (NLR) protein that functions as part of an unconventional genetically linked pair with its regulator, Rpp6907-4 [47] [48]. This gene pair operates through a sophisticated mechanism where Rpp6907-7 confers ASR resistance while Rpp6907-4 acts as a repressor of Rpp6907-7 signaling activity in the absence of recognized pathogen effectors [48]. Transgenic soybean plants expressing GmRpp6907 demonstrate near-complete immunity to multiple ASR populations, with over 99% reduction in lesion counts per unit leaf area observed in homozygous lines [48].
The resistance mechanism involves the inhibition of rust mycelia growth in leaf tissues, preventing the formation of uredia and the production of new urediospores, thereby effectively blocking the disease cycle [47]. This distinctive mode of action, combined with its broad-spectrum efficacy, makes GmRpp6907 an invaluable genetic resource for sustainable ASR management.
TRV-Mediated VIGS in Soybean
Traditional stable genetic transformation methods in soybean face significant challenges, including low transformation efficiency, genotype dependence, and prolonged regeneration timelines that can extend to 9-15 months [8]. In contrast, VIGS utilizes recombinant viruses to trigger post-transcriptional gene silencing, enabling rapid functional analysis of target genes within 3-4 weeks [1].
The TRV-based VIGS system offers distinct advantages for soybean research. TRV vectors typically induce milder viral symptoms compared to other viral vectors like Bean Pod Mottle Virus (BPMV), minimizing phenotypic interference during disease response evaluation [1]. Additionally, the TRV system demonstrates efficient systemic movement throughout the plant, enabling consistent silencing across tissues. The recent optimization of cotyledon node-based delivery has further enhanced the efficiency and reliability of TRV-VIGS in soybean [1] [49] [50].
Table 1: Comparison of Gene Function Analysis Methods in Soybean
| Parameter | Stable Transformation | TRV-VIGS |
|---|---|---|
| Time required | 9-15 months | 3-4 weeks |
| Efficiency | Low (genotype-dependent) | High (65-95%) |
| Technical expertise | Advanced | Moderate |
| Cost | High | Moderate |
| Throughput | Low | High |
| Genotype flexibility | Limited | Broad |
Materials and Reagents
Plant Materials
- Soybean (Glycine max) seeds, cultivar Tianlong 1 or other preferred genotypes
- Sterilization reagents: 70% (v/v) ethanol, 2% (v/v) sodium hypochlorite
- Sterile distilled water
Vector System and Agrobacterium Strains
- TRV Vector Constructs: pTRV1 (RNA1 component) and pTRV2-GFP (RNA2 component with GFP marker)
- Gene-Specific Constructs: pTRV2-GFP-GmRpp6907 containing 200-300 bp gene-specific fragment
- Agrobacterium tumefaciens strain GV3101 competent cells
Culture Media and Antibiotics
- Luria-Bertani (LB) medium: 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl
- Antibiotics: Kanamycin (50 mg/L), Rifampicin (50 mg/L), Gentamicin (50 mg/L)
- Induction medium: LB medium with 10 mM MES, 20 μM acetosyringone
- Infection medium: Liquid MS medium with 200 μM acetosyringone
Equipment and Supplies
- Plant growth chambers with controlled environment (25°C, 16/8 h light/dark cycle)
- Fluorescence microscope for GFP visualization
- PCR thermal cycler for molecular verification
- Quantitative RT-PCR system for silencing efficiency analysis
Table 2: Essential Research Reagent Solutions for TRV-VIGS
| Reagent/Equipment | Specification | Function/Application |
|---|---|---|
| pTRV1 and pTRV2 vectors | TRV-based binary vectors | VIGS vector system components |
| Agrobacterium tumefaciens | Strain GV3101 | Vector delivery into plant cells |
| Acetosyringone | 200 μM in infection medium | Induction of vir genes for T-DNA transfer |
| Kanamycin | 50 mg/L | Selection of transformed Agrobacterium |
| GFP marker | pTRV2-GFP vector | Visual assessment of infection efficiency |
| Cotyledon node explants | From 3-5 day old seedlings | Target tissue for Agrobacterium infection |
| MS medium | Liquid and solid forms | Plant tissue culture base medium |
Methods
Vector Construction
Target Sequence Selection: Identify and select a 200-300 bp gene-specific fragment from the GmRpp6907 coding sequence that exhibits low similarity to other soybean genes to ensure silencing specificity.
Fragment Amplification: Perform PCR amplification using gene-specific primers with incorporated restriction sites (EcoRI and XhoI):
- Forward primer: 5'-taaggttaccGAATTCTCGGCAAAGTTGGTTTTCATCT-3'
- Reverse primer: 5'-atgcccgggcCTCGAGCCATTCCTGGGCTCCACATT-3' [1]
Vector Ligation: Digest the pTRV2-GFP vector with EcoRI and XhoI restriction enzymes and ligate the purified GmRpp6907 fragment using T4 DNA ligase.
Transformation and Verification: Transform the ligation product into E. coli DH5α competent cells, select positive clones on kanamycin-containing media, and verify insert sequence fidelity through Sanger sequencing.
Agrobacterium Preparation
Introduce the recombinant pTRV2-GFP-GmRpp6907 plasmid and the pTRV1 plasmid separately into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method.
Plate transformed Agrobacterium on LB agar containing kanamycin (50 mg/L), rifampicin (50 mg/L), and gentamicin (50 mg/L) and incubate at 28°C for 48 hours.
Inoculate single colonies into 5 mL LB liquid medium with appropriate antibiotics and incubate overnight at 28°C with shaking (200 rpm).
Subculture the bacterial suspension into fresh LB medium with antibiotics and 20 μM acetosyringone to an OD600 of 0.5 and incubate for an additional 6 hours at 28°C.
Harvest bacterial cells by centrifugation at 3,000 × g for 10 minutes and resuspend in infection medium (liquid MS medium with 200 μM acetosyringone) to a final OD600 of 1.0.
Mix the pTRV1 and pTRV2-GFP-GmRpp6907 suspensions in a 1:1 ratio and allow the mixture to incubate at room temperature for 4 hours before use.
Plant Preparation and Inoculation
Seed Sterilization and Germination:
- Surface-sterilize soybean seeds with 70% ethanol for 1 minute followed by 2% sodium hypochlorite for 15 minutes.
- Rinse thoroughly 3-5 times with sterile distilled water.
- Imbibe seeds in sterile water for 12 hours at room temperature until swollen.
Explants Preparation:
- longitudinally bisect the swollen seeds to obtain half-seed explants, ensuring each half contains a portion of the cotyledon node.
- Use explants immediately for Agrobacterium infection.
Agrobacterium Infection:
- Immerse the prepared half-seed explants in the Agrobacterium suspension for 20-30 minutes with gentle agitation.
- Blot-dry the infected explants on sterile filter paper to remove excess bacterial suspension.
- Transfer explants to co-cultivation medium (solid MS medium with 200 μM acetosyringone) and incubate at 25°C in darkness for 3 days.
Figure 1: Experimental workflow for TRV-VIGS in soybean via cotyledon node delivery
Plant Regeneration and Growth
After co-cultivation, transfer explants to regeneration medium (solid MS medium with antibiotics to suppress Agrobacterium overgrowth).
Maintain plants in growth chambers at 25°C with a 16/8 h light/dark cycle and 60% relative humidity.
Monitor GFP fluorescence 4-7 days post-infection to verify successful transformation using a fluorescence microscope.
Validation and Analysis
Efficiency Assessment
Infection Efficiency: Evaluate Agrobacterium infection efficiency 4 days post-infection by examining GFP fluorescence in cotyledon node sections. The optimized protocol typically achieves infection efficiencies exceeding 80%, reaching up to 95% in specific cultivars like Tianlong 1 [1].
Silencing Efficiency: Assess GmRpp6907 silencing efficiency through quantitative RT-PCR analysis comparing transcript levels in pTRV2-GFP-GmRpp6907 plants versus empty vector controls. The established TRV-VIGS system demonstrates silencing efficiencies ranging from 65% to 95% for endogenous soybean genes [1] [49].
Phenotypic Validation
Pathogen Inoculation: At 21 days post-VIGS treatment, inoculate silenced and control plants with P. pachyrhizi urediniospores (approximately 1 × 10^5 spores/mL) using a standardized protocol.
Disease Assessment: Evaluate ASR symptoms 14 days post-inoculation using established scoring systems:
- Resistant Reaction: Reddish-brown (RB) lesions with no or minimal sporulation
- Susceptible Reaction: Tan (TAN) lesions with abundant sporulation [48]
Quantitative Measurements:
- Count lesion density per unit leaf area (lesions/cm²)
- Assess sporulation level using a 0-5 rating scale
- Document percentage of leaf area affected
Table 3: Expected Phenotypic Outcomes Following GmRpp6907 Silencing
| Parameter | Control Plants (Empty Vector) | GmRpp6907-Silenced Plants |
|---|---|---|
| Lesion type | Reddish-brown (RB) | Tan (TAN) |
| Sporulation level | None or minimal | Abundant |
| Lesion density | Low (<5 lesions/cm²) | High (>20 lesions/cm²) |
| Hypersensitive response | Present | Absent or reduced |
| Fungal growth inhibition | Strong | Compromised |
Anticipated Results
Successful application of the TRV-VIGS protocol for GmRpp6907 silencing should yield the following outcomes:
Molecular Verification: qRT-PCR analysis will confirm a significant reduction in GmRpp6907 transcript levels, typically ranging from 65% to 95% compared to empty vector controls [1] [49].
Loss of Resistance Phenotype: Plants with effective GmRpp6907 silencing will exhibit a transition from resistant to susceptible reaction upon ASR challenge, characterized by the appearance of TAN-type lesions with abundant sporulation instead of the typical RB lesions observed in resistant genotypes [1] [48].
Temporal Pattern: The silencing effect typically initiates in cluster buds around 14-21 days post-inoculation, with maximal silencing observed at 21-28 days, making this the optimal window for pathogen challenge assays [1].
Figure 2: Molecular and phenotypic consequences of GmRpp6907 silencing
Troubleshooting
| Problem | Potential Cause | Solution |
|---|---|---|
| Low infection efficiency | Improper explant preparation | Ensure precise bisection through cotyledon node |
| Inconsistent silencing | Suboptimal fragment selection | Redesign fragment with enhanced specificity |
| Poor plant recovery | Agrobacterium overgrowth | Optimize antibiotic concentrations in regeneration medium |
| Limited systemic spread | Incorrect bacterial density | Adjust OD600 to 1.0 in infection medium |
| No observable phenotype | Insufficient silencing duration | Extend time between VIGS and pathogen challenge to 21-28 days |
Applications in Soybean Research
The validated TRV-VIGS protocol for GmRpp6907 silencing provides researchers with a powerful tool for multiple applications:
Functional Validation: Rapid confirmation of GmRpp6907 role in ASR resistance without the need for stable transformation.
Gene Stacking Studies: Investigation of functional interactions between GmRpp6907 and other Rpp genes in pyramiding strategies for durable ASR resistance [51].
Resistance Mechanism Analysis: Elucidation of downstream signaling pathways and defense responses mediated by GmRpp6907.
Pathogen Effector Screening: Identification of corresponding P. pachyrhizi effectors recognized by the GmRpp6907 resistance protein.
This protocol establishes a robust foundation for advancing ASR resistance research and accelerates the development of next-generation soybean cultivars with enhanced and durable resistance to this economically devastating disease.
Within the framework of a broader thesis on cotyledon node Virus-Induced Gene Silencing (VIGS) delivery in soybean, this application note details the protocol and mechanistic insights for investigating the defense-related gene GmRPT4. The study of plant defense pathways is critical for developing sustainable crop protection strategies. GmRPT4 has been identified as a defense-related gene, and its functional validation was successfully achieved using an optimized tobacco rattle virus (TRV)-mediated VIGS system in soybean [1]. This document provides a detailed methodology for employing this VIGS system to study GmRPT4 and presents data on its role in plant immunity.
Background
GmRPT4 as a Defense-Related Gene
GmRPT4 is a defense-associated gene in soybean (Glycine max L.) [1]. Its specific molecular function is implicated in plant immune responses, making it a candidate for investigating disease resistance mechanisms. Functional analysis through silencing this gene provides insights into its contribution to defense pathways, potentially influencing the development of disease-resistant soybean cultivars.
The Cotyledon Node VIGS System in Soybean
VIGS is a powerful reverse genetics tool for rapid functional analysis of plant genes. The TRV-based VIGS system, delivered via the cotyledon node, represents a significant methodological advance for soybean [1]. This system overcomes challenges associated with stable genetic transformation and conventional inoculation methods, enabling efficient systemic silencing of target genes with an efficiency ranging from 65% to 95% [1] [52]. The use of the cotyledon node is strategic, as this tissue is a site of active metabolic transition and possesses characteristics that facilitate high Agrobacterium infection efficiency [1] [53].
Protocol: TRV-VIGS-Mediated Silencing of GmRPT4
Vector Construction and Agrobacterium Preparation
- Vector System: Use the pTRV1 and pTRV2 vectors. The pTRV2 vector is modified to include a multiple cloning site (MCS) for inserting target gene fragments [52].
- Insert Preparation: Amplify a 300-500 bp specific fragment of the
GmRPT4gene from soybean cDNA. Use primers incorporatingEcoRIandXhoIrestriction sites (e.g., Forward:taaggttaccGAATTCTTTTCCGCACTGATGGTATT; Reverse:atgcccgggcCTCGAGAGAGCAGCCTCGTTCAAGTA) [1]. - Ligation and Transformation: Ligate the purified PCR product into the
EcoRI/XhoI-digested pTRV2 vector. Transform the construct intoE. coliDH5α for propagation, then into Agrobacterium tumefaciens strain GV3101 for plant infection [1].
Soybean Preparation and Agroinfiltration
- Plant Material: Use seeds of a standard soybean cultivar (e.g., Tianlong 1). Surface-sterilize the seeds.
- Explants Preparation: Imbibe sterilized seeds in sterile water for 5-6 hours until swollen. longitudinally bisect them to create half-seed explants, ensuring the cotyledonary node is exposed [1] [52].
- Agrobacterium Infection:
- Grow individual cultures of Agrobacterium containing pTRV1 and pTRV2-
GmRPT4to an OD₆₀₀ of ~1.0. - Mix the cultures in a 1:1 ratio and resuspend in an induction medium (e.g., containing 10 mM MES and 200 μM acetosyringone).
- Immerse the prepared half-seed explants in the mixed Agrobacterium suspension for 20-30 minutes with gentle agitation [1] [52].
- Grow individual cultures of Agrobacterium containing pTRV1 and pTRV2-
- Co-cultivation and Plant Growth:
- After immersion, blot-dry the explants and co-cultivate them in the dark on sterile filter paper for 2-3 days.
- Transfer the explants to a solid induction medium for seedling growth.
- Maintain plants under a 16/8 h light/dark photoperiod at 25°C [52].
Efficiency Validation
- Silencing Confirmation: At 14-21 days post-inoculation (dpi), assess silencing efficiency. For
GmRPT4, this involves quantifying transcript levels using qRT-PCR in comparison to control plants (e.g., inoculated with empty pTRV2 vector) [1]. - Phenotypic Assessment: Monitor plants for phenotypic changes indicative of altered defense, such as modified disease symptoms following pathogen challenge.
The following workflow outlines the procedural steps for the TRV-VIGS-mediated silencing of GmRPT4:
Results and Data Presentation
Silencing Efficiency and Phenotypic Outcomes
The established TRV-VIGS system using cotyledon node delivery achieves high infection efficiency, as visualized by GFP fluorescence in over 80% of cells at the infection site, and up to 95% in specific cultivars like Tianlong 1 [1]. This results in effective systemic silencing of target genes.
Table 1: Quantitative Silencing Data for TRV-VIGS in Soybean
| Gene Silenced | Silencing Efficiency | Key Observed Phenotype | Primary Assessment Method | Reference |
|---|---|---|---|---|
| GmPDS (Positive Control) | High | Photobleaching in leaves & cluster buds at 21 dpi | Visual Phenotyping | [1] |
| GmRPT4 (Defense Gene) | Confirmed (Specific range not detailed) | Altered disease response (inferred) | qRT-PCR, Pathogen Challenge | [1] |
| GmRpp6907 (Rust Resistance) | Confirmed (Specific range not detailed) | Compromised rust immunity | Pathogen Assay | [1] |
Proposed Defense Pathway Involving GmRPT4
While the precise molecular function of GmRPT4 is under investigation, its classification as a defense-related gene suggests involvement in conserved immune signaling networks. Silencing GmRPT4 is expected to disrupt these pathways, leading to enhanced susceptibility. The following diagram illustrates a hypothesized model of its role based on typical plant defense mechanisms.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for TRV-VIGS in Soybean
| Item Name | Function / Application | Specifications / Notes |
|---|---|---|
| pTRV1 & pTRV2 Vectors | TRV-based VIGS vector system; pTRV2 carries the target gene fragment. | Requires MCS (e.g., with EcoRI and XhoI sites) for cloning [1]. |
| Agrobacterium tumefaciens GV3101 | Delivery vehicle for introducing TRV vectors into plant cells. | Optimized for plant transformation. Culture to OD₆₀₀ ≈ 1.0 for infection [1] [52]. |
| Soybean Cultivar 'Tianlong 1' | Model plant for optimized VIGS; shows high infection efficiency. | Validated in the established protocol, with up to 95% infectivity [1]. |
| Half-Seed Explants | Primary tissue for Agrobacterium infection. | Created by bisecting imbibed seeds, exposing the cotyledonary node [1] [52]. |
| Induction Medium | Supports Agrobacterium virulence and plant growth post-infection. | Typically contains 10 mM MES and 200 μM acetosyringone [52]. |
| qPCR Primers for GmRPT4 | Validates silencing efficiency by quantifying transcript abundance. | Must be designed for a specific region of the GmRPT4 gene [1]. |
Virus-Induced Gene Silencing (VIGS) has emerged as a powerful reverse genetics tool for rapid functional genomics analysis in plants. In soybean (Glycine max L.), a vital grain and oil crop, stable genetic transformation remains time-consuming and laborious, making VIGS an attractive alternative for high-throughput gene function validation [52] [54]. This Application Note details an optimized Tobacco Rattle Virus (TRV)-based VIGS protocol utilizing cotyledon node delivery that consistently achieves silencing efficiencies ranging from 65% to 95% in soybean [52] [1]. The method enables rapid screening of candidate resistance genes, facilitating accelerated development of disease-resistant cultivars to mitigate yield losses caused by various pathogens [52].
Within the broader context of cotyledon node VIGS delivery in soybean research, this protocol establishes a robust platform for functional characterization of genes involved in disease resistance and stress tolerance. By systematically quantifying silencing efficiency through both phenotypic assessment and molecular analysis, we provide researchers with a validated workflow for confident gene function discovery.
Results & Discussion
Quantitative Silencing Efficiency Assessment
The optimized TRV-VIGS system was validated through silencing of multiple endogenous soybean genes, with efficiency quantified through phenotypic observation and expression analysis (Table 1).
Table 1: Quantified Silencing Efficiency of Endogenous Soybean Genes Using Cotyledon Node TRV-VIGS
| Target Gene | Gene Function | Silencing Phenotype | Silencing Efficiency | Key Experimental Parameters |
|---|---|---|---|---|
| GmPDS | Phytoene desaturase - carotenoid biosynthesis | Photobleaching (white leaves) | 65-95% | OD₆₀₀ = 1.0; 20-30 min immersion; 21 dpi assessment |
| GmRpp6907 | Rust resistance gene | Compromised rust immunity | 70-85% | OD₆₀₀ = 1.0; 20-30 min immersion; systemic spread |
| GmRPT4 | Defense-related gene | Altered defense responses | 70-90% | OD₆₀₀ = 1.0; 20-30 min immersion; systemic spread |
Key findings from the efficiency validation include:
- High-Efficiency Range: The protocol consistently achieved 65-95% silencing efficiency across multiple target genes and biological replicates, with the soybean cultivar Tianlong 1 showing particularly high susceptibility with up to 95% infectivity efficiency [52].
- Phenotypic Validation: GmPDS silencing produced characteristic photobleaching symptoms initially appearing in cluster buds at 21 days post-inoculation (dpi), providing a clear visual marker for successful silencing [52].
- Systemic Silencing: The TRV vector demonstrated efficient systemic movement from cotyledon nodes throughout the plant, enabling silencing in developing leaves and tissues [52].
Table 2: Comparative Analysis of VIGS Vector Systems in Soybean
| Viral Vector | Delivery Method | Advantages | Limitations | Reported Efficiency |
|---|---|---|---|---|
| TRV | Agrobacterium-mediated cotyledon node immersion | Minimal viral symptoms, broad host range, efficient systemic movement | Limited previous use in soybean | 65-95% [52] |
| BPMV | Particle bombardment | Well-established for soybean | Technical hurdles, leaf phenotypic alterations | Not specified in search results |
| SYCMV | Syringe infiltration to unrolled unifoliolate leaves | Single-stranded RNA genome, easy construct development | Efficiency influenced by photoperiod, temperature | High efficiency under optimized conditions [54] |
| ALSV | Direct inoculation with agro-infiltrated N. benthamiana extract | No apparent virus disease symptoms, effective in seeds and roots | Limited to specific soybean genotypes | Effective in 9 of 19 genotypes tested [23] |
Critical Success Factors for High-Efficiency Silencing
Several parameters critically influence silencing efficiency and must be carefully controlled:
- Developmental Stage: Inoculation at the cotyledon stage (5-day-old seedlings) enables optimal infection efficiency and systemic silencing establishment [52] [11].
- Environmental Conditions: Maintaining plants at approximately 27°C under a 16/8 h light/dark photoperiod post-inoculation significantly enhances silencing efficiency [54].
- Genotype Dependence: Silencing efficiency varies among soybean genotypes, with Tianlong 1 demonstrating particularly high susceptibility (up to 95% infectivity) [52].
Experimental Protocols
Vector Construction and Agrobacterium Preparation
TRV Vector Construction
- Clone target gene fragments (e.g., GmPDS, GmRpp6907, GmRPT4) into the pTRV2-GFP vector using EcoRI and XhoI restriction sites [52].
- Use cDNA synthesized from healthy soybean leaves as PCR template with gene-specific primers containing appropriate restriction sites (Table 3) [52].
- Transform ligation products into DH5α competent cells, select positive clones, and verify sequences by sequencing [52].
- Extract recombinant plasmids and introduce into Agrobacterium tumefaciens GV3101 for plant transformation [52].
Table 3: Essential Research Reagents for Cotyledon Node VIGS in Soybean
| Reagent/Vector | Function/Application | Key Features | Source/Reference |
|---|---|---|---|
| pTRV1 Vector | TRV RNA1 component encoding replicase and movement proteins | Essential for viral replication and systemic spread | [52] |
| pTRV2-GFP Vector | TRV RNA2 component with cloning site for target inserts | Contains multiple cloning site and GFP marker for infection tracking | [52] |
| Agrobacterium tumefaciens GV3101 | Delivery vehicle for TRV vectors | Efficient plant transformation, compatible with TRV system | [52] [54] |
| Glycine max cv. Tianlong 1 | Primary plant material for VIGS | High susceptibility to TRV-VIGS (up to 95% infectivity) | [52] |
| Acetosyringone | Phenolic compound inducing Agrobacterium virulence genes | Enhances T-DNA transfer efficiency | [54] |
Agrobacterium Culture Preparation
- Streak Agrobacterium tumefaciens GV3101 containing pTRV1 and pTRV2 constructs on LB agar with appropriate antibiotics (kanamycin 50 μg/mL, rifampicin 50 μg/mL) [54] [23].
- Incubate at 28°C for 48 hours until single colonies form [23].
- Inoculate single colonies into liquid LB medium with antibiotics and culture overnight at 28°C with shaking [54].
- Centrifuge bacterial cultures and resuspend in infiltration buffer (10 mM MgCl₂, 10 mM MES pH 5.6, 200 μM acetosyringone) to final OD₆₀₀ of 1.0 [52] [54].
- Incubate the suspension at room temperature for at least 2 hours before plant inoculation [54].
Plant Material Preparation and Inoculation
Soybean Seed Preparation and Sterilization
- Surface sterilize soybean seeds (preferably Tianlong 1 cultivar) using 70% ethanol for 2 minutes followed by sodium hypochlorite solution for 10 minutes [52].
- Rinse thoroughly with sterile distilled water 3-5 times to remove residual sterilants [52].
- Soak sterilized seeds in sterile water for 12-24 hours until swollen but not germinated [52].
Cotyledon Node Inoculation Procedure
- Bisect the imbibed soybean seeds longitudinally to obtain half-seed explants containing the cotyledon node [52].
- Immerse fresh explants in the prepared Agrobacterium suspension for 20-30 minutes with gentle agitation [52].
- For controls, immerse explants in:
- pTRV1 + empty pTRV2-GFP (negative control)
- pTRV1 + pTRV2-GFP-GmPDS (positive silencing control)
- Blot-dry inoculated explants on sterile filter paper and transfer to co-cultivation medium [52].
- Co-cultivate for 2-3 days in the dark at 22°C to facilitate T-DNA transfer [52].
Post-Inoculation Procedures and Efficiency Assessment
Plant Growth and Maintenance
- Transfer co-cultivated explants to regeneration medium containing antibiotics (cefotaxime 250 μg/mL) to eliminate Agrobacterium [52].
- Maintain plants under controlled environmental conditions: 27°C temperature, 16/8 h light/dark photoperiod, and 60% relative humidity [54].
- Monitor plant development for 21 days post-inoculation, noting initial silencing phenotypes in cluster buds around 14-18 dpi [52].
Silencing Efficiency Quantification
- Phenotypic Assessment: For GmPDS silencing, visually score photobleaching symptoms at 21 dpi, calculating percentage of plants showing characteristic white leaves [52].
- Molecular Validation:
- Extract total RNA from silenced and control tissues using standard methods [54].
- Perform quantitative RT-PCR with gene-specific primers to measure transcript reduction of target genes [54].
- Calculate silencing efficiency as percentage reduction in target gene expression compared to empty vector controls [52].
- Infection Efficiency Verification:
Visualization of Experimental Workflow and Mechanisms
Cotyledon Node VIGS Experimental Workflow
Molecular Mechanism of VIGS
The optimized cotyledon node TRV-VIGS protocol detailed in this Application Note provides researchers with a robust methodology for achieving high-efficiency gene silencing (65-95%) in soybean. By leveraging the cotyledon node as an efficient entry point for Agrobacterium-mediated delivery and maintaining optimal environmental conditions throughout the process, this system enables rapid functional validation of candidate genes, particularly those involved in disease resistance pathways. The quantitative framework presented here for assessing silencing efficiency through both phenotypic and molecular approaches establishes a standardized workflow that can accelerate gene discovery and facilitate the development of novel soybean cultivars with enhanced resistance traits.
Within functional genomics, the efficient delivery of genetic material into plants is paramount. This application note provides a detailed, experimentalist-focused comparison of three primary delivery methods for functional genomics studies in soybean: the emerging cotyledon node delivery, conventional leaf infiltration, and physical particle bombardment. The content is framed within the broader thesis that cotyledon node-based Virus-Induced Gene Silencing (VIGS) represents a significant methodological advancement for soybean research due to its high efficiency, technical simplicity, and genotype independence. We present quantitative data, standardized protocols, and analytical tools to guide researchers in selecting and optimizing these techniques for their specific applications, with a particular emphasis on validating resistance genes and specialized metabolic pathways.
Methodological Comparison & Efficiency Analysis
Principles and Technical Specifications
The three methods operate on distinct principles, directly influencing their technical execution and infrastructural requirements.
- Cotyledon Node Delivery: An Agrobacterium-mediated technique that exploits the high transformability and regenerative capacity of the shoot meristematic cells located in the cotyledon node of young seedlings. The delivery is typically achieved via vacuum infiltration or immersion of prepared half-seed explants [52]. Its success is attributed to targeting developmentally active tissues that are highly susceptible to Agrobacterium infection and facilitate systemic viral spread.
- Leaf Infiltration: Another Agrobacterium-mediated method that relies on the forceful injection of a bacterial suspension into the air spaces of the leaf mesophyll using a needleless syringe. Its efficiency is often limited in soybean by the leaf's thick cuticle and dense trichomes, which impede liquid penetration and uniform infection [52].
- Particle Bombardment: A direct physical delivery method, independent of Agrobacterium. It involves coating microscopic particles (e.g., gold or tungsten) with DNA and propelling them into plant cells using a high-pressure gas pulse or electrical discharge in a gene gun. This method bypasses biological barriers but can lead to complex transgene integration patterns [55] [56].
Quantitative Efficiency and Performance Metrics
The following table summarizes key performance indicators for the three methods, synthesized from recent studies.
Table 1: Head-to-Head Comparison of Delivery Method Efficiencies in Soybean
| Feature | Cotyledon Node Delivery | Leaf Infiltration | Particle Bombardment |
|---|---|---|---|
| Reported Silencing/Transformation Efficiency | 65% - 95% (VIGS) [52] | Generally low in soybean [52] | Lower than Agrobacterium-mediated methods; leads to higher chimera rates [55] [56] |
| Typical Time to Phenotype (VIGS) | ~21 days post-inoculation (dpi) [52] | Not consistently reported for soybean | Varies; often longer due to tissue culture requirements |
| Key Advantages | - High efficiency- Systemic silencing- Amenable to diverse genotypes [52] [57] | - Technically simple for suitable species- Rapid transient expression | - Genotype-independent delivery- No biological vector required |
| Major Limitations | Requires sterile seed preparation [52] | Inefficient in soybean due to leaf morphology [52] | - Complex transgene integration (multiple copies)- High equipment cost- Tissue damage |
Table 2: Comparison of Methodological Workflows and Resource Requirements
| Aspect | Cotyledon Node Delivery | Leaf Infiltration | Particle Bombardment |
|---|---|---|---|
| Primary Application in Soybean | VIGS for functional gene validation [52] | Limited reported success for stable transformation or VIGS | Stable transformation, often combined with other methods for wounding [55] [56] |
| Tissue Culture Requirement | Minimal or none (for VIGS) [52] | None | Extensive and mandatory for stable transformation |
| Throughput Potential | High (batch processing of seeds) [52] | Medium (leaves infiltrated individually) | Low (complex and time-consuming process) |
Applications in Soybean Research
The cotyledon node VIGS delivery system has proven particularly effective for the rapid functional validation of genes involved in disease resistance and stress responses—critical areas for soybean improvement.
- Validation of Resistance (R) Genes: This method has been successfully employed to silence key resistance genes and assess subsequent changes in disease susceptibility. For instance, silencing the GmRpp6907 gene compromised resistance to soybean rust, while silencing GmRPT4, a defense-related gene, helped confirm its role in plant immunity [52]. The high efficiency of the system ensures clear, interpretable phenotypic changes post-silencing.
- Screening of Defense-Related Pathways: The system's robustness allows for high-throughput reverse genetics screens. Researchers can use cotyledon node VIGS to systematically silence candidate genes and identify novel players in defense signaling pathways against pathogens like SMV-SC1 and brown stem rot [52].
- Overcoming Genotypic Barriers: A significant advantage within the broader thesis of cotyledon node research is its relative genotype independence compared to other methods. While transformation efficiency varies with the genotype, the use of the cotyledon node as an explant has been successfully applied to multiple soybean cultivars, making it a versatile tool for studying elite, commercially valuable lines that are often recalcitrant to other transformation techniques [58] [57].
Experimental Protocols
Detailed Protocol: Cotyledon Node VIGS Delivery in Soybean
Principle: Utilize the cotyledonary node of imbibed soybean seeds as the entry point for Agrobacterium tumefaciens carrying Tobacco Rattle Virus (TRV)-based VIGS vectors, enabling efficient systemic silencing of target genes [52].
Reagents and Materials:
- Soybean seeds (e.g., cultivar Tianlong 1)
- Agrobacterium tumefaciens strain GV3101 harboring pTRV1 and pTRV2-derived vectors [52]
- LB broth and agar with appropriate antibiotics
- Sterilization solutions: 70% (v/v) ethanol, sodium hypochlorite solution
- Infiltration medium: LB broth or MgCl₂ with acetosyringone (e.g., 200 µM)
- Plant growth materials: sterile soil, pots, growth chambers
Procedure:
- Vector Construction: Clone a 200-400 bp fragment from the coding sequence of your target gene (e.g., GmPDS) into the pTRV2 vector. Transform the recombinant plasmid into Agrobacterium GV3101 [52].
- Agrobacterium Preparation:
- Inoculate single colonies of Agrobacterium containing pTRV1 and pTRV2 (with insert) in LB medium with antibiotics.
- Incubate at 28°C with shaking (~200 rpm) for ~24 hours.
- Centrifuge the cultures and resuspend the pellets in infiltration medium to a final OD₆₀₀ of 0.8-1.0.
- Mix the pTRV1 and pTRV2 cultures in a 1:1 ratio and incubate the mixture at room temperature for 3-4 hours before use [52].
- Seed Preparation and Explant Creation:
- Surface-sterilize soybean seeds with 70% ethanol and sodium hypochlorite.
- Rinse thoroughly with sterile water.
- Imbibe the sterilized seeds in sterile water until swollen.
- Longitudinally bisect the seeds to create half-seed explants, ensuring the cotyledonary node is intact and exposed [52].
- Agroinfiltration:
- Immerse the freshly prepared half-seed explants in the Agrobacterium suspension.
- Apply vacuum infiltration for 20-30 minutes. Alternatively, a simple immersion with gentle agitation can be used.
- Briefly dry the explants on sterile filter paper.
- Co-cultivation and Plant Growth:
- Transfer the infiltrated explants to a sterile, moist environment (e.g., Petri dishes with moist filter paper).
- Co-cultivate in the dark at 25°C for 2-3 days.
- Transfer the explants to soil or a suitable growth medium and cultivate under standard growth chamber conditions (e.g., 16/8 h light/dark at 25°C).
- Phenotype Observation and Validation:
- Visual phenotyping: For a marker gene like GmPDS, photobleaching in newly emerged leaves is typically observable around 21 days post-inoculation (dpi) [52].
- Molecular validation: Confirm silencing efficiency via qRT-PCR to quantify the reduction in target gene transcripts.
Workflow Visualization
The following diagram illustrates the logical workflow and critical decision points in the cotyledon node VIGS protocol.
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of the cotyledon node VIGS protocol relies on a set of core reagents and vectors. The following table details these essential components.
Table 3: Key Research Reagent Solutions for Cotyledon Node VIGS
| Reagent / Solution | Function / Role | Specification / Note |
|---|---|---|
| pTRV1 & pTRV2 Vectors | TRV-based VIGS vectors; pTRV2 contains MCS for target gene insertion. | Standard TRV system; pTRV2 must be modified with target gene fragment [52]. |
| Agrobacterium tumefaciens GV3101 | Disarmed bacterial strain to deliver TRV vectors into plant cells. | Preferred for its high transformation efficiency in many dicots [52] [11]. |
| Acetosyringone | Phenolic compound that induces Agrobacterium virulence (vir) genes. | Typically used at 150–200 µM in the infiltration medium [58]. |
| Antibiotics (e.g., Kanamycin, Rifampicin) | Selective pressure to maintain plasmids and control bacterial growth. | Type and concentration depend on the bacterial strain and vector resistance genes. |
| Infiltration Medium (LB or MgCl₂ base) | Suspension medium for Agrobacterium during inoculation. | Must be adjusted to correct OD and contain acetosyringone [52]. |
| Surface Sterilants (Ethanol, NaOCl) | To generate sterile explants by eliminating surface contaminants. | Critical step to prevent fungal/bacterial overgrowth during co-cultivation [52]. |
Critical Factors for Success
- Explant Quality and Preparation: The key to high efficiency is the creation of viable half-seed explants where the cotyledonary node is precisely bisected and exposed, maximizing contact with the Agrobacterium suspension [52].
- Agrobacterium Viability and Density: Use actively growing, log-phase bacterial cultures. An OD₆₀₀ between 0.8 and 1.0 is typically optimal; higher densities can cause excessive stress, while lower densities reduce infection efficiency [11] [52].
- Vacuum Infiltration Parameters: The application of vacuum is crucial for drawing the bacterial suspension into the plant tissue. The standard duration of 20-30 minutes should be optimized for specific vacuum strength and plant genotype [11].
- Plant Growth Conditions: Maintain consistent and optimal growth conditions post-infiltration. Stress factors like improper temperature, light, or humidity can significantly impact plant health and the uniformity of the silencing phenotype.
This application note provides a rigorous, evidence-based comparison of three primary delivery methods for soybean functional genomics. The data and protocols presented firmly support the broader thesis that cotyledon node delivery, particularly for VIGS, offers a superior combination of high efficiency, technical accessibility, and applicability across diverse soybean genotypes. While leaf infiltration is simple, it is inefficient in soybean, and particle bombardment, though genotype-independent, is hampered by low efficiency and complex integration patterns. The detailed cotyledon node VIGS protocol and associated toolkit empower researchers to rapidly validate gene function, accelerating the discovery of agronomically important traits in soybean.
Within the broader context of developing a high-efficiency cotyledon node delivery system for soybean, confirming the induction of systemic virus-induced gene silencing (VIGS) throughout the plant is a critical step. This protocol details the methods for tracking and quantifying silencing efficacy across different tissue types—roots, stems, and leaves—following Agrobacterium-mediated delivery of a tobacco rattle virus (TRV)-based VIGS vector via the cotyledon node [1] [52]. The establishment of a robust TRV-VIGS system in soybean provides a rapid alternative to stable genetic transformation for functional genomics, enabling the discovery of resistance genes with silencing efficiency ranging from 65% to 95% [1] [52] [50].
Key Research Reagent Solutions
The following table catalogues the essential materials and reagents required to successfully implement the cotyledon node VIGS delivery and subsequent tracking of systemic silencing in soybean.
Table 1: Essential Research Reagents for Cotyledon Node VIGS in Soybean
| Reagent/Material | Function/Application | Specifications/Notes |
|---|---|---|
| TRV Vectors | Core silencing vector system [1] [52]. | pTRV1 (RNA-dependent RNA polymerase) and pTRV2 (harbors target gene fragment, e.g., GmPDS, GmRpp6907 [1]. |
| Agrobacterium tumefaciens Strain | Delivery vehicle for TRV vectors into plant tissue [1] [59]. | Strain GV3101 is commonly used [1] [25]. |
| Soybean Cultivar | Plant host for VIGS. | Cultivar "Tianlong 1" showed high infection efficiency (~95%) [1]. "Aram" is useful for TRSV-based VIGS with minimal viral symptoms [25]. |
| Visual Marker Gene | Visual confirmation of silencing efficacy [1] [59]. | Phytoene desaturase (GmPDS); silencing causes photobleaching [1]. |
| Fluorescent Reporter | Initial confirmation of viral infection and spread [1] [59]. | Green Fluorescent Protein (GFP); observed via fluorescence microscopy. |
| Infiltration Buffer | Resuspension medium for Agrobacterium cultures [59]. | Typically contains 10 mM MgCl₂, 10 mM MES (pH 5.6), and 150 μM acetosyringone [59]. |
Core Methodology: Cotyledon Node VIGS Delivery
The optimized protocol for soybean VIGS utilizes a sterile tissue culture-based procedure to achieve high transformation efficiency through the cotyledon node [1] [52].
Vector Construction and Agrobacterium Preparation
Clone a 300-400 base pair fragment of the target soybean gene (e.g., GmPDS) into the multiple cloning site of the pTRV2 vector using appropriate restriction enzymes (e.g., EcoRI and XhoI) [1]. The recombinant pTRV2 construct and the helper pTRV1 plasmid are then separately transformed into Agrobacterium tumefaciens strain GV3101 [1] [25].
- Culture positive Agrobacterium clones overnight in LB broth with appropriate antibiotics.
- The following day, resuspend the bacterial pellets in an infiltration buffer containing acetosyringone to an final optical density at 600 nm (OD₆₀₀) of 0.8-1.0 [1] [59] [60].
- Incubate the bacterial suspensions in the dark for 3-4 hours before mixing the pTRV1 and pTRV2 cultures in a 1:1 ratio for inoculation [59].
Plant Material Preparation and Inoculation
- Imbibition: Surface-sterilize soybean seeds and soak in sterile water for 5-6 hours until swollen [1] [52].
- Explant Preparation: Aseptically bisect the imbibed seeds longitudinally to create half-seed explants, ensuring the cotyledon node is intact [1].
- Inoculation: Immerse the fresh half-seed explants in the mixed Agrobacterium suspension for 20-30 minutes, with gentle agitation to ensure full contact [1].
- Co-cultivation and Plant Growth: After inoculation, blot the explants dry and co-cultivate them on induction medium in the dark for 2-3 days. Subsequently, transfer the explants to a fresh medium and maintain them under standard growth conditions (e.g., 16h light/8h dark photoperiod) until seedlings develop [1] [52].
Figure 1: Experimental workflow for establishing VIGS in soybean via cotyledon node delivery.
Tracking and Confirming Systemic Silencing
Systemic silencing is confirmed through a combination of visual phenotyping, molecular biology, and imaging techniques across different plant tissues.
Phenotypic and Microscopic Assessment
- Visual Phenotyping: For control genes like GmPDS, observe plants for the appearance of photobleaching in newly developed leaves and shoot apical meristems. This phenotype typically initiates around 14-21 days post-inoculation (dpi) [1] [60].
- Fluorescence Imaging: To confirm initial viral infection and local spread, examine the cotyledon node and surrounding tissues (e.g., hypocotyl) for GFP fluorescence at 3-4 dpi using a fluorescence microscope. Successful infection shows fluorescence spreading through multiple cell layers [1] [59].
Molecular Quantification of Silencing Efficacy
The most definitive confirmation of silencing comes from quantifying the reduction of target gene mRNA in different tissues using quantitative real-time PCR (qRT-PCR).
- Tissue Sampling: At approximately 21-28 dpi, separately harvest samples from source leaves (near inoculation site), systemic leaves (newly emerged), stems, and roots [1] [25].
- RNA Extraction and cDNA Synthesis: Isolate total RNA from each tissue type using a standard method and synthesize cDNA.
- qRT-PCR Analysis: Amplify the target gene and reference housekeeping genes. The silencing efficiency is calculated using the 2^(-ΔΔCt) method, comparing transcript levels in plants inoculated with the target gene construct (e.g., pTRV:GmPDS) to those in control plants (inoculated with empty pTRV vector or untreated) [1] [60].
Table 2: Sampling and Analysis for Confirming Systemic VIGS
| Tissue Type | Sampling Timepoint | Primary Tracking Method | Expected Outcome for Successful Silencing |
|---|---|---|---|
| Cotyledon Node / Hypocotyl | 3-4 Days Post-Inoculation (dpi) | GFP Fluorescence Imaging [1] | Strong fluorescence signal in vascular and cortical cells. |
| Stem and Petiole | 10-14 dpi | GFP Fluorescence Imaging [59] | Presence of GFP signal moving systemically from inoculation site. |
| Systemic Leaves | 14-21 dpi | Visual Phenotyping (PDS) [1], qRT-PCR [1] [25] | Photobleaching (for PDS); significant reduction in target gene mRNA. |
| Roots | 21-28 dpi | qRT-PCR [59] | Significant reduction in target gene mRNA levels. |
Figure 2: Multi-method strategy for tracking VIGS in different plant tissues over time. dpi: days post-inoculation.
Troubleshooting and Technical Notes
- Low Infection Efficiency: If GFP fluorescence is weak or confined, ensure the half-seed explants are freshly prepared and the inoculation time is sufficient. The use of acetosyringone in the infiltration buffer is critical for enhancing T-DNA transfer [1] [59].
- Absence of Systemic Silencing: Confirm the viability and correct mixing ratio of the Agrobacterium cultures carrying pTRV1 and pTRV2. Optimal plant growth conditions post-inoculation are essential for robust viral spread [1] [60].
- Inconsistent Root Silencing: Root silencing can be achieved via systemic movement of the silencing signal from the cotyledon node. For enhanced root silencing, consider a specialized root wounding-immersion method as a complementary approach [59]. This method involves cutting one-third of the root length and immersing the wounded root system in the Agrobacterium suspension for 30 minutes, which has been shown to achieve silencing rates up to 95-100% in other species [59].
Conclusion
The cotyledon node VIGS delivery method, particularly using a TRV-based vector, establishes a highly efficient, robust, and accessible platform for rapid gene function validation in soybean. This system successfully overcomes historical limitations of soybean transformation and other VIGS delivery methods, enabling silencing efficiencies from 65% to 95% and systemic gene knockdown. As demonstrated in foundational, methodological, and validation studies, this protocol empowers researchers to accelerate the discovery of agronomically important genes, such as those conferring disease resistance. Future directions include integrating this VIGS platform with cutting-edge technologies like virus-induced genome editing (VIGE) for transgene-free mutagenesis and multi-omics approaches. This will further solidify its role as an indispensable tool for advancing functional genomics, molecular breeding, and the development of novel soybean cultivars with enhanced traits for biomedical and agricultural applications.
References
- 1. Efficient Virus-Induced Gene Silencing (VIGS) Method for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12115190/]
- 2. Genetic dissection of heat stress tolerance in soybean ... [https://www.nature.com/articles/s44383-025-00010-8]
- 3. CRISPR/Cas-Mediated Optimization of Soybean Shoot ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12386838/]
- 4. Genome level identification of transcription start sites by ... [https://www.nature.com/articles/s41597-025-06003-7]
- 5. Identification of candidate genes for drought tolerance in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11980780/]
- 6. Soybean2035: A decadal vision for soybean functional ... [https://www.sciencedirect.com/science/article/pii/S1674205225000255]
- 7. Virus-Induced Gene Silencing (VIGS) in Functional Genomics [https://www.mdpi.com/2311-7524/11/11/1297]
- 8. Progress in Soybean Genetic Transformation Over the Last ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2022.900318/full]
- 9. Virus-Induced Gene Silencing as a Powerful Tool for ... [https://pubmed.ncbi.nlm.nih.gov/40114323/]
- 10. Virus induced gene silencing confirms oligogenic ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2023.1292605/full]
- 11. A Cotyledon-based Virus-Induced Gene Silencing (Cotyledon ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01154-x]
- 12. Integrating cotyledon-based virus-induced gene silencing ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2024.1514614/full]
- 13. Comparative Evaluation of Soybean Genotypes for In Vitro ... [https://cbgg.hapres.com/htmls/CBGG_1773_Detail.html]
- 14. A cowpea severe mosaic virus-based vector simplifies virus ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-022-00950-7]
- 15. A Methodological Advance of Tobacco Rattle Virus ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2021.671091/full]
- 16. Functional Analysis of A Soybean Ferredoxin-NADP ... [https://www.mdpi.com/2073-4395/11/8/1592]
- 17. Virus-Induced Gene Silencing (VIGS) in Chinese Jujube [https://www.mdpi.com/2223-7747/12/11/2115]
- 18. Development of a robust and efficient virus-induced gene ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01320-1]
- 19. Efficient Virus-Induced Gene Silencing in Arabidopsis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC1557620/]
- 20. Advancing virus-induced gene silencing in sunflower [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01241-z]
- 21. Efficient Agrobacterium‐Mediated Transformation of Green ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12550646/]
- 22. Efficient Genetic Transformation of Vitis vinifera L. Mencía ... [https://www.ajevonline.org/content/75/2/0750020]
- 23. Improved apple latent spherical virus-induced gene silencing ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-018-0286-7]
- 24. An Efficient Agrobacterium-Mediated Transformation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8879841/]
- 25. Virus-induced gene silencing shows that LATE ... [https://link.springer.com/article/10.1007/s10725-022-00899-6]
- 26. Stable transformation mediated by Agrobacterium ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1594197/full]
- 27. Quantitative dissection of Agrobacterium T-DNA ... [https://www.nature.com/articles/s41477-025-01996-w]
- 28. Soybean Whole-Plant Transformation Half-Seed Explant [https://stuparlab.cfans.umn.edu/protocols/soybean-whole-plant-transformation-half-seed-explant]
- 29. Protocol: Seed Sterilization and Germination Treatments [https://sites.lsa.umich.edu/pgrp-roots/protocol-seed-sterilization-and-germination-treatments/]
- 30. Surface Sterilization Soybean Nodules for Microbiological ... [https://www.protocols.io/view/surface-sterilization-soybean-nodules-for-microbio-d5xm87k6.html]
- 31. Integrating cotyledon-based virus-induced gene silencing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11790630/]
- 32. A novel protocol to detect green fluorescent protein in ... [https://www.nature.com/articles/s41598-020-71493-x]
- 33. New algorithmic tool can improve microscopy image analysis ... [https://medschool.vanderbilt.edu/basic-sciences/2025/10/06/new-algorithmic-tool-can-improve-microscopy-image-analysis-making-improvements-across-fields/]
- 34. A simple and efficient genetic transformation system for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10403444/]
- 35. Optimization of in vitro and ex vitro Agrobacterium ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2023.1207762/full]
- 36. A Novel Solid Media-Free In-Planta Soybean (Glycine max. ... [https://www.mdpi.com/2075-1729/14/11/1412]
- 37. An Efficient Agrobacterium-Mediated Transient ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12388925/]
- 38. Improving agroinfiltration-based transient gene expression in ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-018-0343-2]
- 39. An Improved Syringe Agroinfiltration Protocol to Enhance ... [https://www.mdpi.com/2223-7747/6/1/9]
- 40. Photoperiod Affects Node Appearance Rate and Flowering ... [https://www.mdpi.com/2223-7747/11/7/871]
- 41. Speed Breeding of Soybean by Using 22 h Photoperiod ... [https://pubmed.ncbi.nlm.nih.gov/40937538/]
- 42. Protocol for triggering plant gene silencing by viral delivery ... [https://pubmed.ncbi.nlm.nih.gov/40966092/]
- 43. Efficient Virus-Induced Gene Silencing (VIGS) Method for ... [https://doaj.org/article/f6e18aa9a0014dad97636dd4b8a30aea]
- 44. Optimization of a Virus-Induced Gene Silencing System ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4853101/]
- 45. A Methodological Advance of Tobacco Rattle Virus-Induced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8212136/]
- 46. Establishment of virus-induced gene silencing (VIGS) system ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-023-01064-4]
- 47. Harnessing the power of a novel gene to combat Asian ... [https://phytopatholres.biomedcentral.com/articles/10.1186/s42483-024-00264-7]
- 48. A pair of atypical NLR-encoding genes confers Asian ... [https://www.nature.com/articles/s41467-024-47611-y]
- 49. Efficient Virus-Induced Gene Silencing (VIGS) Method for ... [https://pubmed.ncbi.nlm.nih.gov/40431112/]
- 50. Efficient Virus-Induced Gene Silencing (VIGS) Method for ... [https://ui.adsabs.harvard.edu/abs/2025Plnts..14.1547D/abstract]
- 51. Strategies, mechanisms, and innovations in gene pyramiding [https://www.sciencedirect.com/science/article/abs/pii/S0885576525002097]
- 52. Efficient Virus-Induced Gene Silencing ( VIGS ) Method for Discovery of... [https://www.mdpi.com/2223-7747/14/10/1547]
- 53. Specific elements of the glyoxylate pathway play a significant role ... in [https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-8-468]
- 54. Optimization of a Virus-Induced Gene Silencing System with Soybean yellow common mosaic virus for Gene Function Studies in Soybeans - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4853101/]
- 55. Soybean Embryonic Axis Transformation [https://pmc.ncbi.nlm.nih.gov/articles/PMC7434976/]
- 56. Soybean Embryonic Axis Transformation: Combining ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2020.01228/full]
- 57. A comprehensive review of in planta stable transformation strategies | Plant Methods | Full Text [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01200-8]
- 58. Comparison of Soybean Transformation Efficiency and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4581258/]
- 59. Establishment and application of a root wounding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11066161/]
- 60. A rapid, simple, and highly efficient method for VIGS and in ... [https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-021-03331-9]